The dog that didn't bark
It looks like even symptomatic SARS-CoV-2 infections in the U.S. dates back at least to November 2019......
https://brownstone.org/articles/evidence-of-early-spread-in-the-us-what-we-know/
In the article “The Dog that Didn’t Bark”, The Brownstone Institute mentioned a new study from the American Red Cross that indicated that “above-specificity-limit” levels of SARS-CoV-2 antibodies have been found in blood donors across the United States, as early as in December 2020.
Unlike in Europe and in South America where samples were saved for retrospective testing for a long period of time, allowing the retrospective discovery of SARS-CoV-2 infections as early as September 2019 at high statistical confidence, Biological samples in the United States are very rarely saved for a period longer than a week or banked for retrospective analysis unless the institution itself has some specific research programs that have taken and stored biological samples—by chance. This renders the retrospective screening of samples from patients for SARS-CoV-2 infections nearly impossible within the U.S, although some notable exceptions exist such as a Boston study developing a SARS-CoV-2 specific antigen assay accidentally finding evidence of such antigens in sick patients dating back as early as October 2019.
This unfortunately means that the molecular detection of pre-pandemic SARS-CoV-2 circulation would have to be mainly through the serological testing of patients that were sick before 2020, did not travel, and were no longer sick afterward, as well as from any samples that just happened to be taken right after the indication of “strange pneumonia cases” in Wuhan, but before the official first importation of SARS-CoV-2 into the United States.
Even with little sample left from the pre-pandemic period, some notable detections that satisfied the above conditions have been found.
Are there other symptomatic SARS-CoV-2 infections in USA-WA before WA1?
Before the official date of importation of USA-WA-1 (if it was imported at all, as opposed to the result of community transmission), on 27/12/2019, “two days after Christmas”, “Jean, a 64-year-old retired nurse, suffered through a series of worsening symptoms: a dry, hacking cough, a fever and body aches, and finally, a wheeze that rattled her lungs.”. A chest X-ray taken at the time of hospital admission indicates pneumonia, while the patient slowly improved across two medical visits after treatment with Duoneb, a medication that is usually used to treat Asthma. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5537120/ However, it should also be mentioned that the two specific drugs in Duoneb, Ipratropium bromide, and salbutamol, are also frequently used to treat ARDS (Acute respiratory distress syndrome), which the drugs helps with improvement by facilitating fluid clearance from the lungs. Almost the soon as antibody tests become available, this patient decided to take a SARS-CoV-2 antibody test, which comes up positive. The patient did not travel outside the United States, indicating that limited community transmission in WA may have already begun in December 2019.
Santa Clara: The earliest official COVID-19 deaths obtained by autopsies don’t have travel history.
During an effort of retrospectively testing autopsy cases in Santa Clara, California, two patients, neither having a travel history, that have died on Feb. 6 and Feb. 17, were found to be positive for SARS-CoV-2 infection, two weeks before the first officially confirmed COVID-19 death in the United States on 29 February 2020, in Kirkland, Washington.
This early detection of SARS-CoV-2 infection in these deaths illustrated one of the reasons why the United States was so bad at tracing SARS-CoV-2 spread in the community prior to March 2020: “Both of the Santa Clara County deaths occurred as the Centers for Disease Control and Prevention had tightly restricted tests to those who displayed respiratory symptoms and recently traveled to China or had close contact with an infected person. Cody said local officials often had to call the CDC and discuss the specifics of individual cases before the agency would grant permission for testing.” Community cases and any cases that may have predated any known importations from China were simply excluded from testing.
Alabama: First case requiring ECMO, five days before the first death in Wuhan
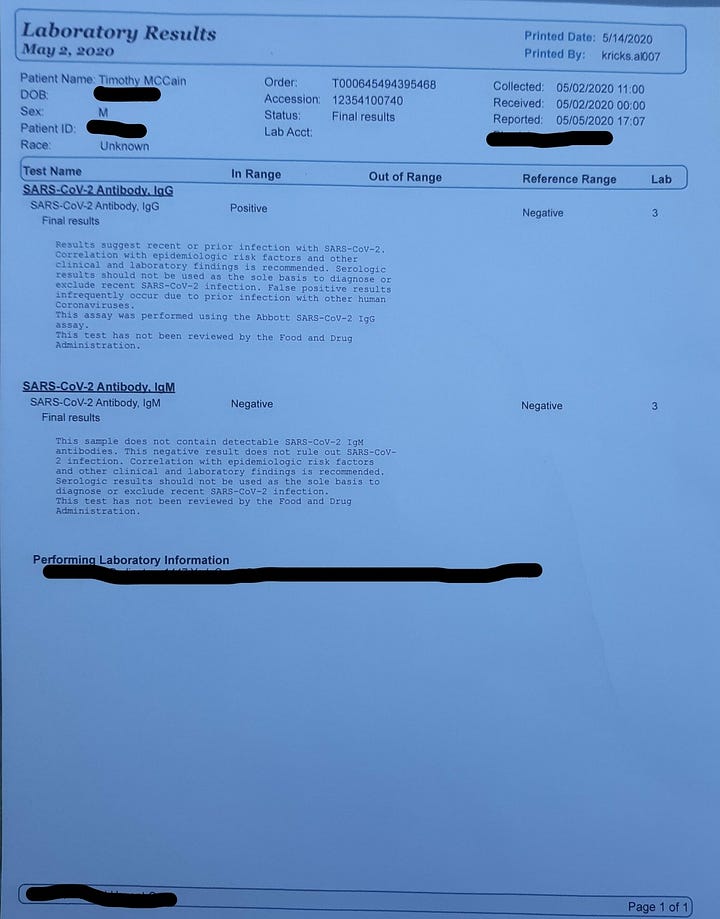
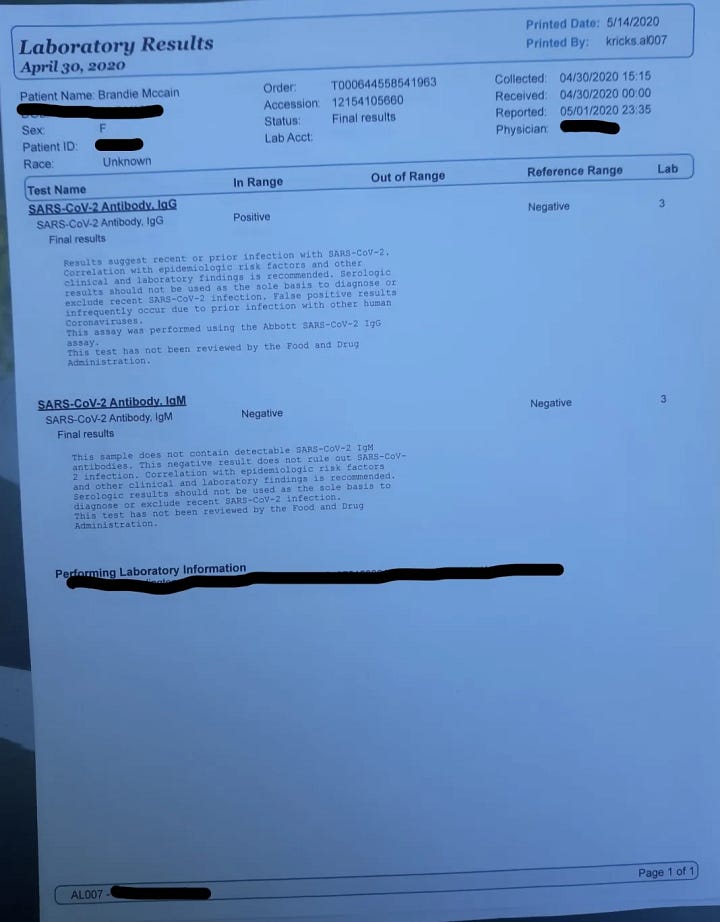
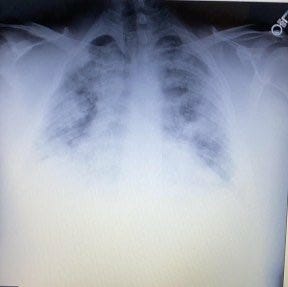

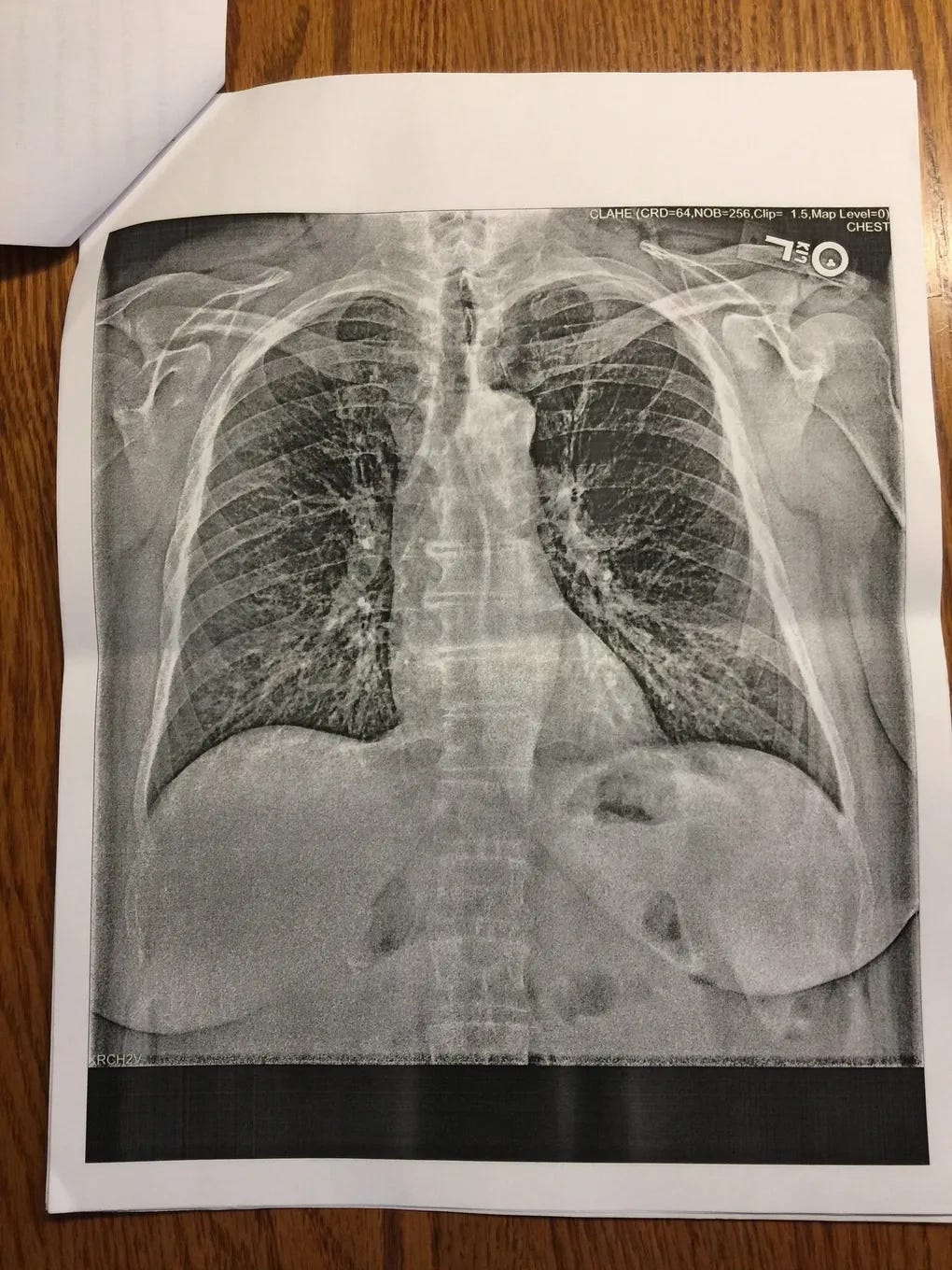
Let’s describe in the official Wuhan language for this family cluster in Alabama: 39F having symptoms onset on 15/12/2019. Husband, 38M, onset “a few days later”. Family members began to show symptoms on 26/12/2019. The last travel history within the cluster was a “Caribbean cruise”, attended by 39F, happening during 2-10/11/2019. The closest community contact of 38M was in Montgomery, where he travels to work. There is no recent contact or travel history to China. 38M is found to be positive for coinfection of IAV, whereas 39F tested negative for influenza despite the same symptoms experienced by both. IgG test turned positive on both at end of April 2020, and no IgM was found. ECMO use was initiated on 04/01/2020 when the Lungs were found to be entirely white on chest X-ray. One of the possible causes for the rapidly deteriorating condition of 38M may have been an early prescription of a “steroid shot” and a “steroid pack” of pills, before the phase where supplemental oxygen therapy begins to be initiated. As it has been found in randomized controlled trials that administration of glucocorticoids such as dexamethasone, leads to rapid deterioration of conditions for the patients (observed as a lower and rapidly worsening PaO2/FiO2 ratio) and an increase in mortality when the patient does not require supplemental oxygen or respiratory support, this early misuse of corticosteroids in 38M may have led to the resultant exaggeration of symptoms and eventual ECMO use.
Florida: It looks like the positive antibody test found there were specific……
At least 11 residents in a neighborhood two blocks in size in Delray Beach, Florida, reported symptoms compatible with symptomatic COVID-19 in late November-early December 2019. All residents tested positive for antibodies as the test became available. All these residents “haven't been sick since”, despite there is now significant community spread as of Mar-Apr 2020 in Florida, indicating that these antibodies are likely already present during the beginning of the officially announced epidemic at this location. This gives significant weight to the possibility that the positive antibody tests found in these residents were connected to their last major respiratory illness, the mysterious outbreak that sickened 11 within a small neighborhood (of just 2 blocks) in Florida.
Kansas: death certificates revised to “COVID-19 pneumonia“ as early as the first death in Wuhan
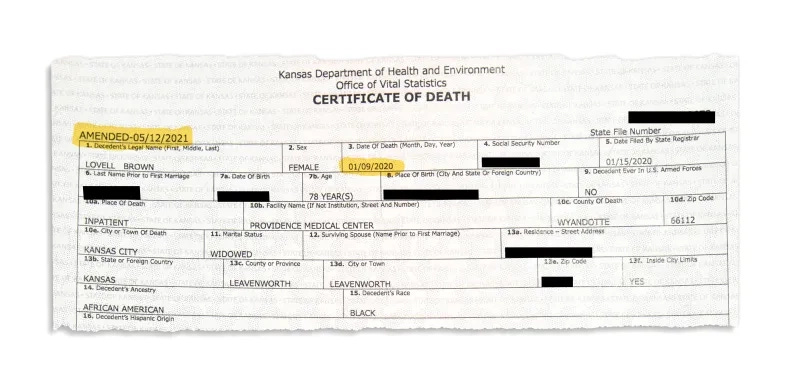
During one of the many revisions of death certificates as autopsies were being performed on dead bodies, one of the death certificates for a woman in Kansas, which experienced fatigue symptoms with loss of taste in Christmas of 2019, and died in an ICU on 09/01/2020, was amended to “COVID-19 pneumonia”. While the exact detail behind this revision is still shrouded in mystery, this could have been linked to samples that were saved during autopsy shortly before the cremation of the body, as the ICU that performed the revision has yet to produce any details on the reason for this revision. This woman also lacked any travel history, which could indicate community transmission being the cause, driving the first date of importation for the military town of Leavenworth to at or even before the 11 December 2019 onset date of the first case at the Huanan market.
New York: Positive PCR the soon as they had fresh samples?
https://www.nature.com/articles/s41467-021-23688-7
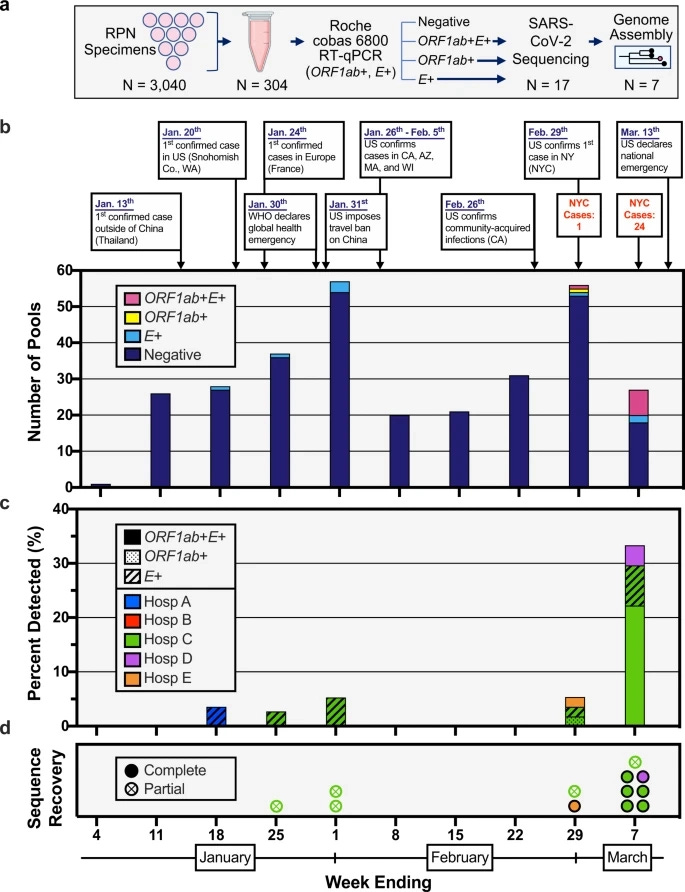
While a lack of archived samples has considerably hampered the search for early evidence of SARS-CoV-2 infection in the United States, efforts that began “immediately following the 8 January 2020 CDC Health Alert Network advisory regarding the outbreak of pneumonia of unknown etiology, later identified as COVID-19.” begun collecting samples starting in 11/01/2020, and then weekly, in 5 hospitals within NYC. Samples, which were usually stored at 4 degrees C for no more than a week (which sadly means that there were no archived human samples from the city of New York—but not a complete absence of all archived samples from the state of New York), were collected, archived and then tested using E- and RdRp- specific PCR for evidence of SARS-CoV-2 RNA in pools of 10. Positive samples were then subjected to next-generation sequencing to deduce the SARS-CoV-2 genomes found within these samples. From the very first pool of samples that were archived immediately after collection and a negative respiratory pathogen PCR test (as opposed to upwards of 4 days in storage at 4 degrees C, as in the 11/01/2020 pool), which are the sample pools taken on 18/01/2020, positive tests of the E protein CDS began to show in the sample pools that have been tested. Comparable percentages of positive samples in all samples were collected across the 3 weeks from 18/01/2020 to 01/02/2020, indicating that relatively stable, cryptic circulation has taken place in this era. Next-generation sequencing recovered “partial genomes” in one library on 25/01/2020 and in two libraries on 01/02/2020.
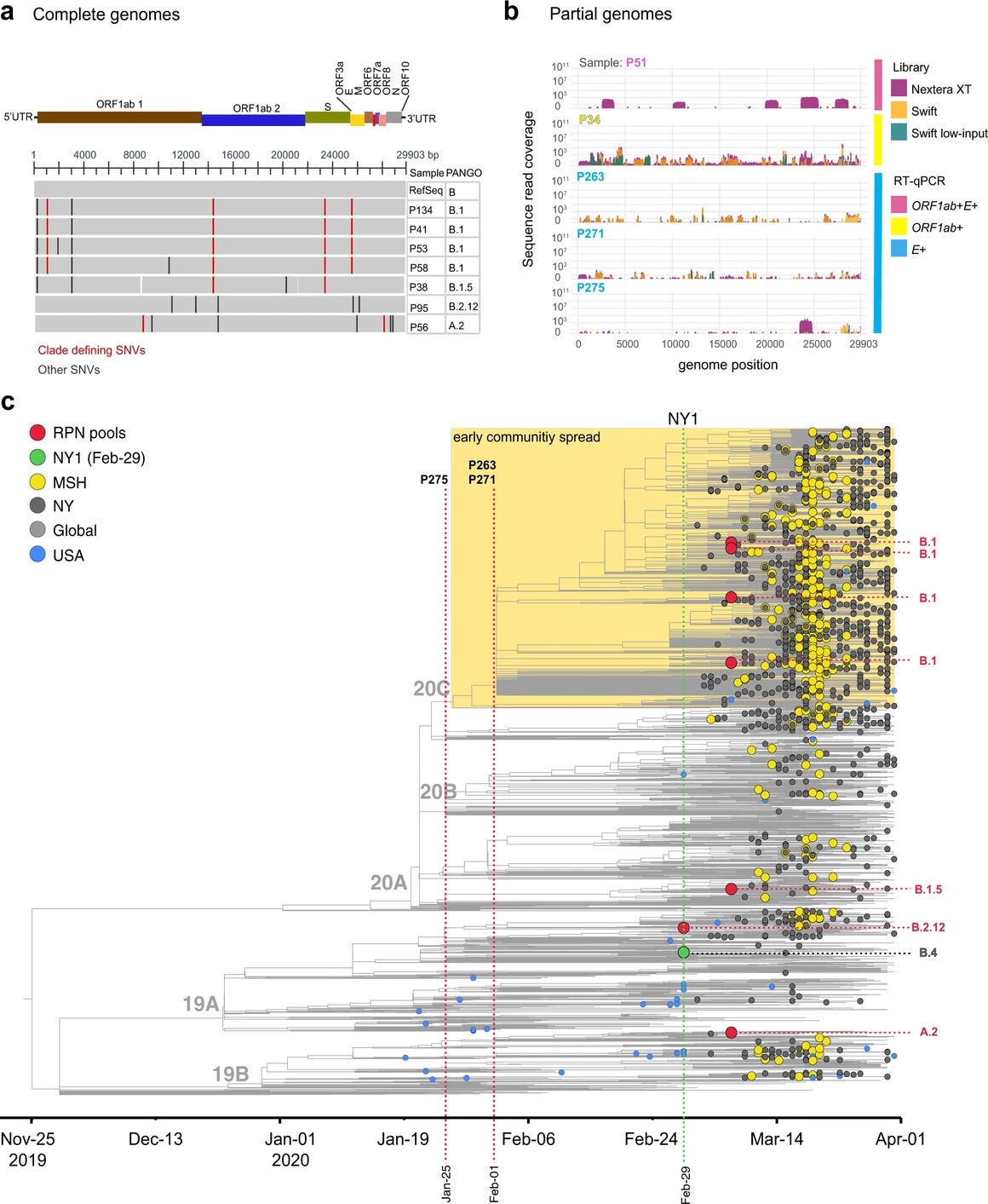
Examination of their obtained sequences deposited under PRJNA717974 indicated that all samples have been filtered so that only SARS-CoV-2 reads were reported. Only samples where they indicated the “partial recovery of SARS-CoV-2 genome” were deposited. By examining the short sequencing reads within PRJNA717974, the earliest sample they give on their SRA (25-01-2020) was found to belong to some “B lineage of SARS-CoV-2” given the 28144T observed in the reads. Mutations 28254Del, 29301C, 29213-29214 TT->GA/partial, 28547G/minor(4 reads), 28405G/minor(4 reads), 28360G/minor(11 reads), 28000T, and 889G were found within this sample pool at varying fractions, indicating multiple sources of the SARS-CoV-2 genome within this library. While this pool is sampled at about the same time the first B.1 sequences were officially sampled in Europe, reads that escape (one A/C mismatch at A28352C corresponding to a T(ref)->G(alt) alignment at the negative strand and likely pre-Wuhan origin (3 reads), amongst others including mismatches at the 3' end of the N1 primer) the N1-R primer has been found. Unlike C→U and A→G transitions, which retain a wobble base pair between the primer and the mutated template and are usually tolerated by qRT-PCR, transversions create kinks and bulges within the primer-template duplex and cause substantial reduction of the Tm of the primer-template duplex, preventing proper primer binding especially when the annealing temperature is set close to the designed Tm of the qRT-PCR primers to ensure specificity. This indicates that strains that were invisible to the N-specific CDC PCR may have circulated in this period.
Mutations like 28000T and 28254Del are found only in the B.1 lineage on NextStrain. This indicates that B.1 is most likely already there on 25/01/2020. In one of the 01/02/2020 pools, 23403 is found to be G, whereas, in the other 01/02/2020 pool, 241 is found to be T. This indicates that a B.1 derivative is present in USA-NY as early as in 01/02/2020.
Nucleotide positions 9400-9425 and 12094-12119 (the 2 human lung-specific MiRNA binding sites) were not found in the 25/01/2020 or 01/02/2020 pools.
Onondaga: Positive deer as early as January 05?
https://www.pnas.org/doi/10.1073/pnas.2114828118
While obtaining retrospective human samples for testing of SARS-CoV-2 is difficult in the U.S. due to how biological samples from patients are handled within this country, this is not true for the obtaining of retrospective animal samples—which were archived for as long as a decade. In January 2020, a sampling of white-tailed deer across 4 states, USA-MI, USA-NY, USA-PA, and USA-IL revealed three samples positive for SARS-CoV-2 neutralizing antibodies—in a trend that steadily rises across the years 2019-2021. All 3 2020 samples were taken in Onondaga, where 1/4 samples taken on 05/01/2020 and 2/14 samples taken on 28/01/2020 were found to be positive for SARS-CoV-2 neutralizing antibodies. As it is found that the B.1-specific mutation Spike D614G is required for the infection of free-ranging white-tailed deer and transmission between deer, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8833191/table/t01/
(All deer-derived SARS-CoV-2 lineages belong to B.1.)
https://www.researchgate.net/publication/349981365_Susceptibility_of_White-Tailed_Deer_Odocoileus_virginianus_to_SARS-CoV-2
(The tiger B.1 D614G strain was used) https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8382122/
(When WA1 and B.1.1.7 was used together, transmit only B.1.1.7.)
https://www.nature.com/articles/s41586-021-04353-x
(B.1.2, B.1.582, and B.1.596 were found.)
This finding is consistent with the observation that early strains found in human samples circulating within NYC at the very beginning of human clinical sample banking were derivatives of the B.1 lineage. As spillover into deer in Onondaga would have to happen between 19/12/2019 and 05/01/2020 in order for the first SARS-CoV-2 neutralizing antibody positive sample within them to be found in 05/01/2020, this puts the date of genesis for the B.1 lineage to be at least as early as the earliest known official cases within Wuhan—considering that the B.1 lineage was not observed in Wuhan or China until March 2020, this necessitates the presence of an index case for USA-NY well before the first case of SARS-CoV-2 infection at the Huanan seafood market, and very likely to be as early as November 2019.
As Onondaga is 288 miles away from New York City, with the city of Albany in between, Community transmission that generate sufficient number of infections and spread sufficiently far to begin infecting deer in Onondaga, or to build up SARS-CoV-2 Titres within wastewater biosolids high enough for transmission to deer from Biosolids application to become possible, would have taken two to three weeks, in addition to the 14 days that were observed in inoculated deer from infection for antibodies that reacted to the RBD protein of SARS-CoV-2 (which the sVNT test targeted, as the inhibition of SARS-CoV-2 RBD binding to ACE2 require specifically antibodies that reacted to the SARS-CoV-2 RBD, as opposed to general inhibition of live virus which could happen on other epitopes not necessarily on the Spike, that begin to show at 7 Dpi). This would place the index case within NYC to be at least as early as 09/12/2019, three days before the symptom onset of the first HSM case at 11/12/2019.
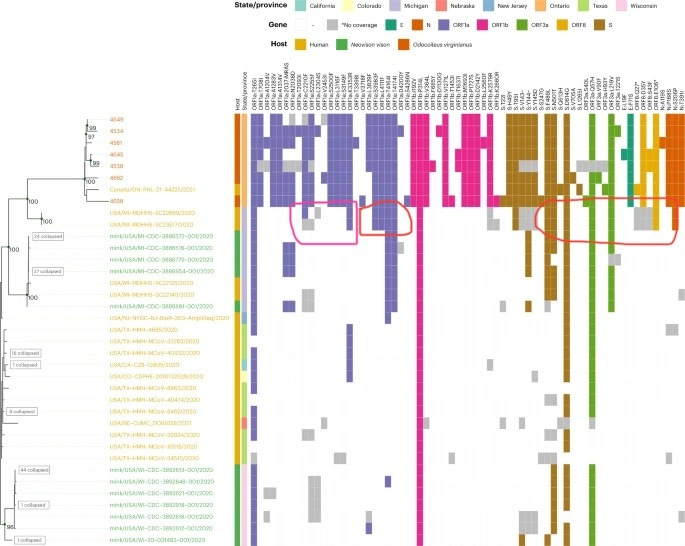
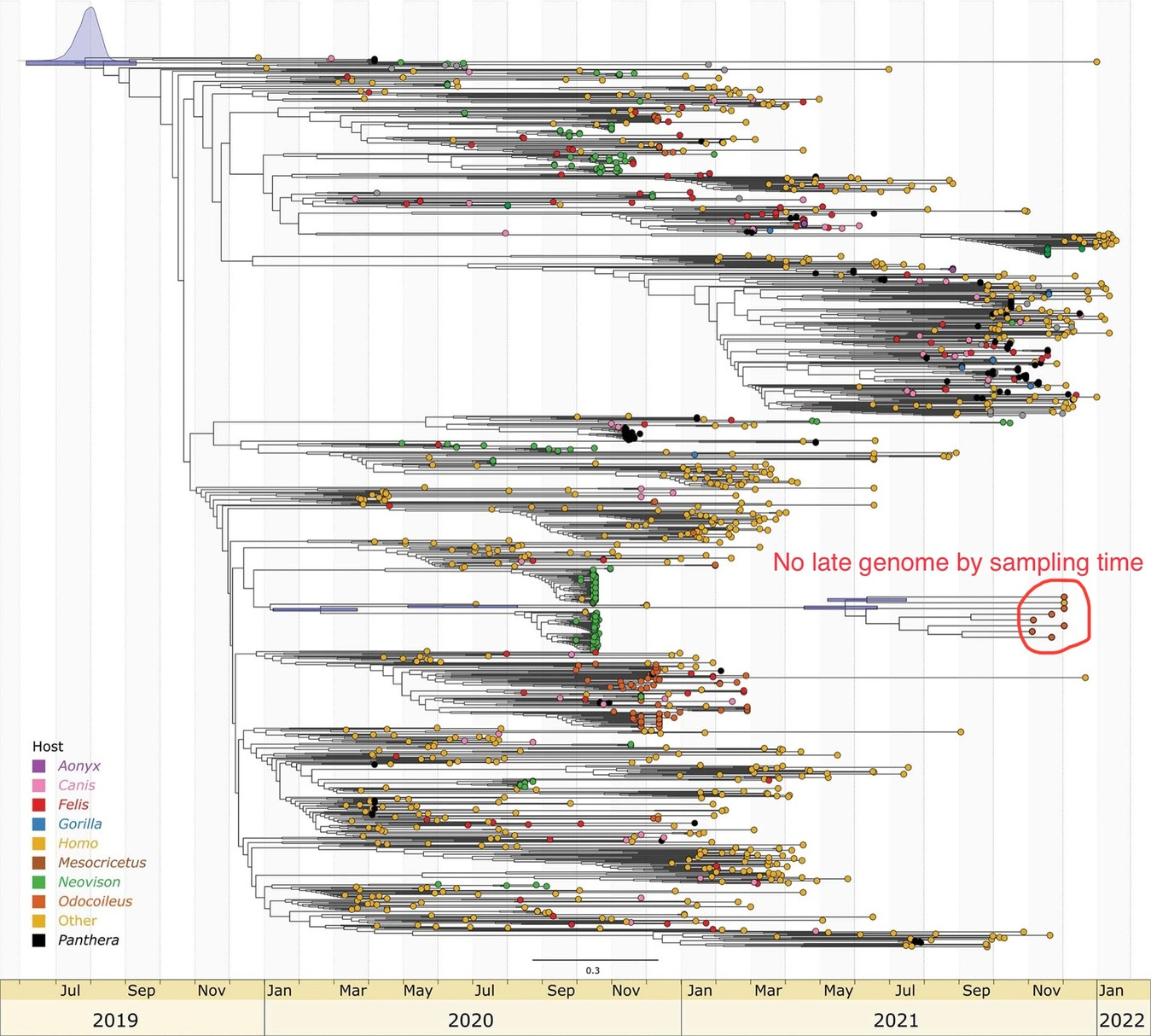
More deer surprise: Most divergent ORF1ab known…… In Canadian Deer?
https://www.nature.com/articles/s41564-022-01268-9
While serological survey places the index case of B.1 within North America to comparable or even before the official index case within Wuhan, Phylogenetic analysis of samples obtained from white-tailed deer in Ontario Canada places the tMRCA between this hypermutated branch of SARS-CoV-2 to the rest of the SARS-CoV-2 phylogenetic tree to as early as July-September 2019—While these samples were claimed to cluster with mink and human isolates within USA-MI, examination of the mutations found within these deer-derived genomes revealed that this similarity is largely due to two recombinant regions between ORF1ab AA 2210-4714 (where discordant positions K3353R, T4614I and T4714I were found, where isolates containing only K3353R and isolates containing only T4614I/T4714I were found in the branch leading to B.1.641 indicating recombination at this position) and after S AA 486 (where an unusually low number of RBD mutations were observed despite a large number of NTD mutations). These two recombinant regions included the E and N protein CDS, which may have enabled the detection of this highly divergent lineage through conventional qRT-PCR assays but only after the recombination have taken place.
While the B.1.641 lineage is found to be more divergent than even Omicron at time of sampling, and with divergence covering a range that is comparable to a Variant-Of-Concern (VOC) clade, the sampling time of this cluster happened too close to each other for the evolutionary rate within this particular clade to be determined to any useful accuracy. While the article claimed that the B.1.641 lineage shared a tMRCA with the rest of B.1 “between May and August 2020”, it was then revealed that this is based not on an internally determined mutation rate within the B.1.641 lineage itself, but “on the basis of a root-to-tip regression of the global phylogeny” and were based on the sampling time-constrained global phylogeny of SARS-CoV-2 without considering recombination. This renders the article’s proclaimed tMRCA circular and a support for it using evolutionary rates internally determined within the clade lacking.
Unlike Omicron where there may have been positive selection pressure on the Spike protein, despite being even more divergent than Omicron in terms of phylogeny, “the ORF1ab analysis identified significant relaxation of selection amongst the white-tailed deer lineage (P = 0.0032). These signatures of neutrality were further supported by the even distribution of conserved mutations in proportion to gene/product length (Extended Data Fig. 2).” for lineage B.1.641, Indicating that selection pressure cannot be the cause of the observed high divergence of B.1.641, especially on the non-recombinant region (ORF1ab) of the B.1.641 backbone.
An analysis of the mutational spectrum of SARS-CoV-2 in deer, humans, and mink revealed an equally unremarkable pattern, where an overrepresentation of G→U, C→U, and A→G is obtained, and that the range of mutations seen in deer is not unseen in humans. As free-ranging white-tailed deer are not exposed to known RNA mutagens that are found in several drugs that are only taken by humans, chemical mutagenesis can be ruled out as the cause of this unexpected level of divergence. As B.1.641 was sampled about the same time as the most divergent Omicron sequence taken in their phylogenetic tree analysis, with no justifiable mechanism for the hypermutation of B.1.641, and assuming the fastest observed rate of divergence for circulating SARS-CoV-2 genomes (Omicron), the tMRCA between B.1.641 and the rest of the SARS-CoV-2 phylogenetic tree (on the nonrecombinant regions) could be 1.14 to 1.27 times longer than the time of divergence between MN908947.3 and the most divergent Omicron sequence used in the phylogenetic reconstruction of this article, giving a time of divergence for the nonrecombinant regions of B.1.641 to as early as July-September 2019—comparable to that of the first respiratory outbreak in Fairfax county, Virginia.








Thanks for this. I've been researching "early spread" for 2 1/2 years and you are the first person who has even mentioned many of the same presumed cases I have. For example, you highlight the cases of Tim and Brandie McCain of Sylaucauga, Alabama. I sent my exclusive feature story on the McCains to about 30 news organizations and none would run it ... until finally Tracy Beanz at uncoverDC.com ran it.
The info on the deers and the case where the birth certificate was later revised were not known to me. Good luck with your research. I don't think many of us think "early spread" is a big deal, but it's actually a huge deal.
you missed the best part of the latest deer paper:
https://pubmed.ncbi.nlm.nih.gov/36357713/
no immune response!
https://youtu.be/tTsOn-4DLmM?t=2238