Study of the USA-WA-1 case report and CFR pattern in USA-WA reveal critical biological lacuna of the Chinese NNDRS system
Was the most recent common ancestor of SARS-CoV-2 too mild to register on China's surveillance system?
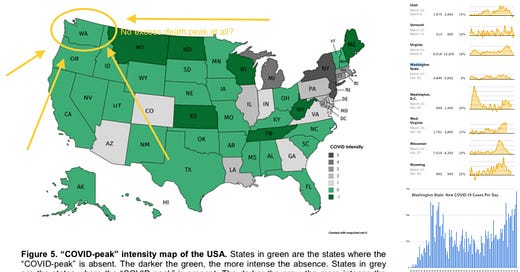
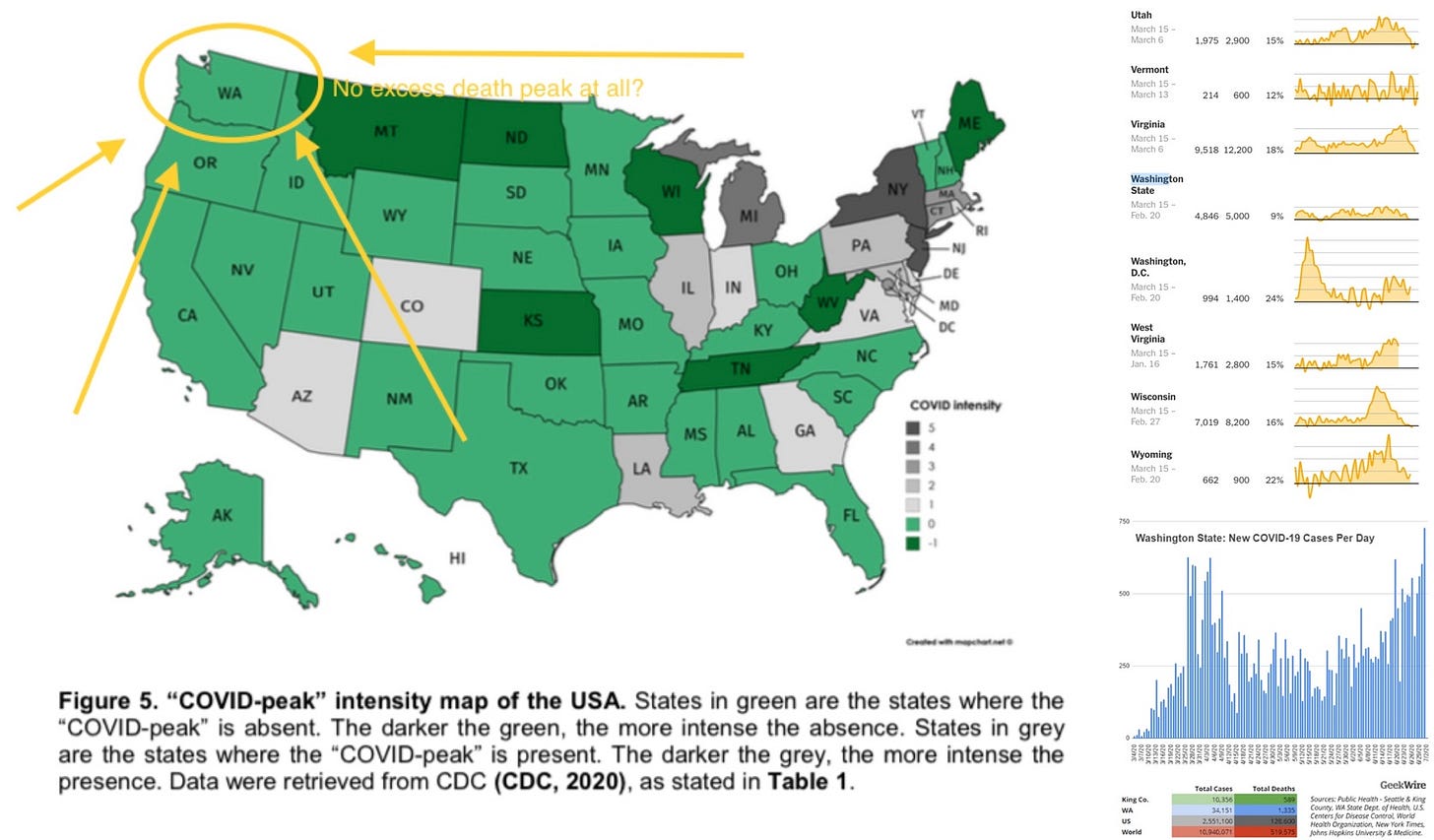
Recently, a French study of the “COVID peak” across the United States has generated more questions than it has answered.

While Washington (USA-WA) was the location where the first official US COVID-19 case was detected, mass testing revealed hundreds of cases every day as the measure began in 03/2020, the excess death curve in USA-WA had flatlined. At the same time, locations in the U.S. that received an introduction of lineage B have seen a massive spike in deaths.
https://www.nytimes.com/interactive/2021/01/14/us/covid-19-death-toll.html
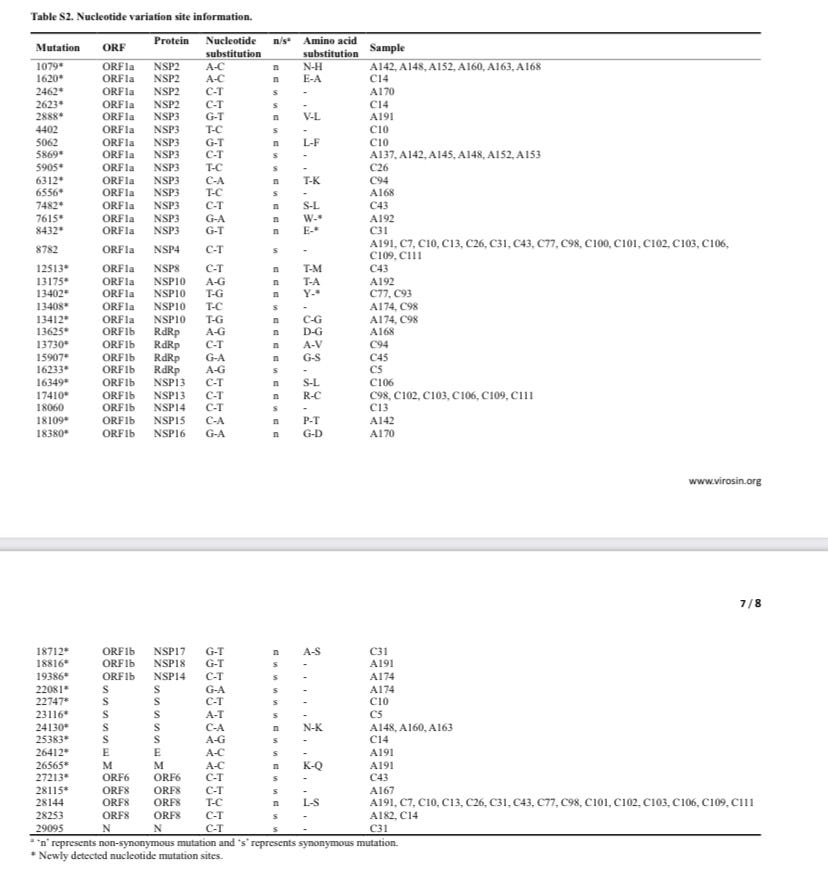
The cause of this could be two-fold. It is possible that WA1 is extremely mild, to the point that the Pneumonia of Unknown Etiology (PUE) standards used in the Chinese National Notable Disease Reporting System (NNDRS) or any of the case definition standards that were active in Wuhan prior to the lockdown, will be unlikely met with an infection of this strain. This is evidenced by the fact that the eventual discovery in Wuhan of this strain would be from one of the mass testing efforts during the lockdown era on 26/01/2020 from a throat swab sample collected from “patients admitted to various Wuhan health care facilities”, hCoV-19/Wuhan/0126-C13/2020 https://pubmed.ncbi.nlm.nih.gov/33851337 (Note: all GWH BioSample GPS coordinates given by this article are the same, and were defaulted to the geometric center of downtown Wuhan.).
This is a week after the presentation of the WA1 patient on 19/01/2020.
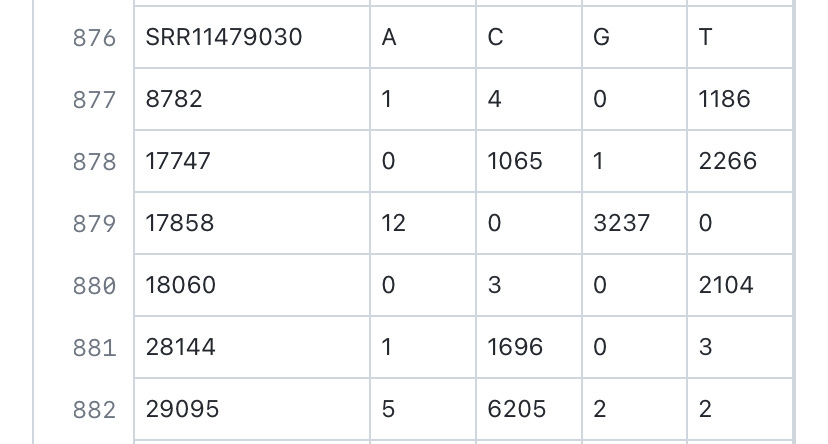
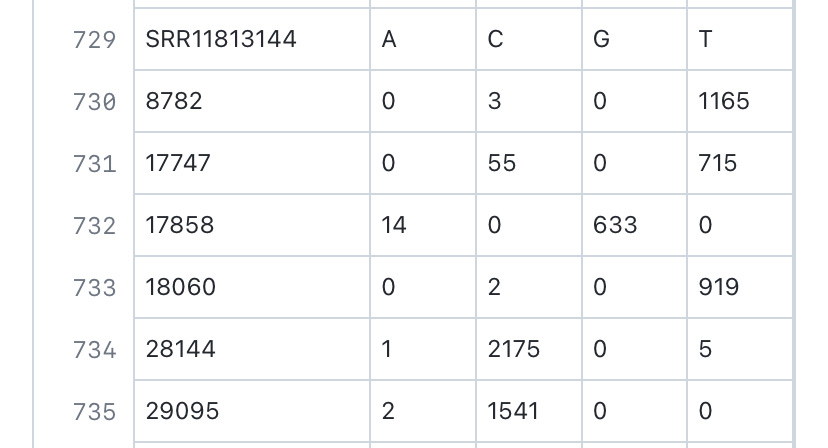
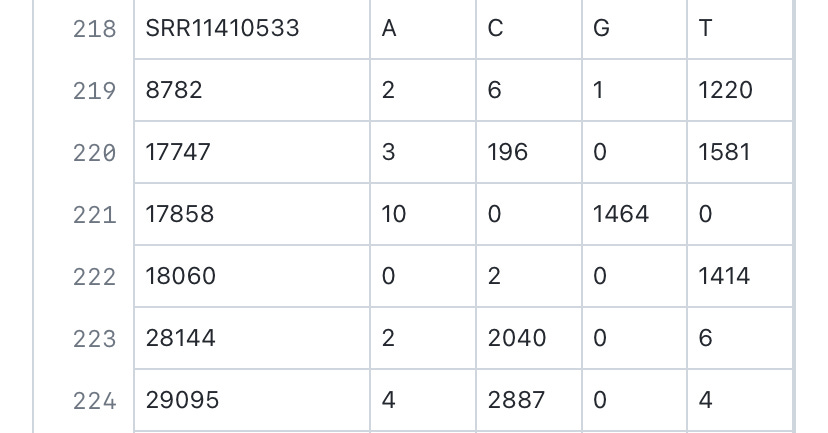
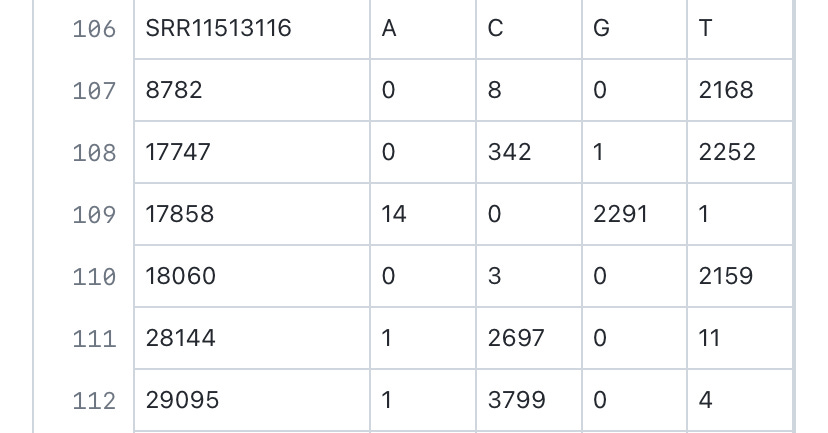
https://ngdc.cncb.ac.cn/bioproject/browse/PRJCA002539
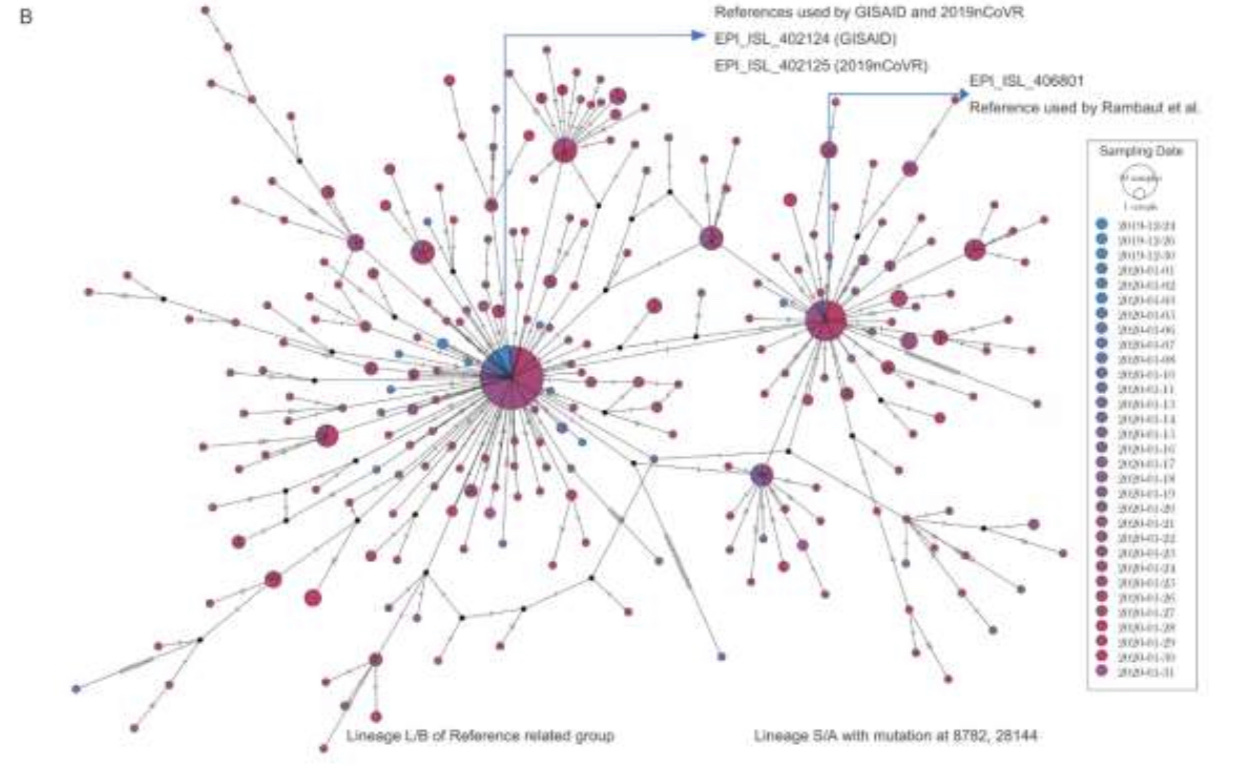
Interestingly, while samples that were labeled “0125-ANNN” comes with the age and sex of the patients, and contained mainly lineage B sequences of SARS-CoV-2, samples that were labeled as “0126-CNNN” comes with no information on the age and sex of the patients, and contained mainly lineage A sequences of SARS-CoV-2. This may indicate that while the samples “0125-ANNN” were taken from hospitalized inpatients that usually provide such detailed records, the samples “0126-CNNN” were taken from non-hospitalized outpatients (that have been admitted to clinics, but not hospitalized) where records are less detailed and age/sex data is often not provided when these patients were sampled by the WIV for this particular study.
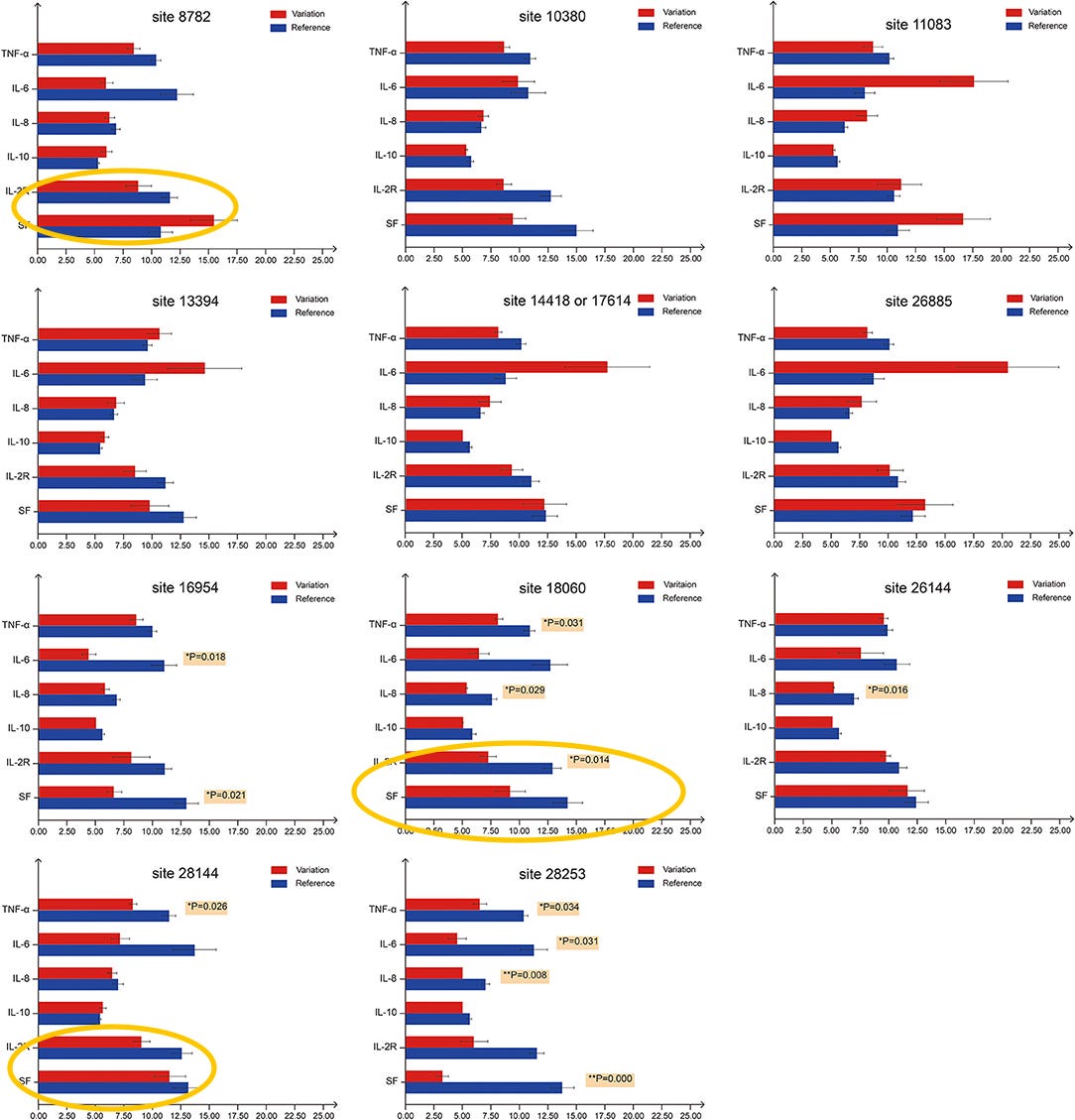
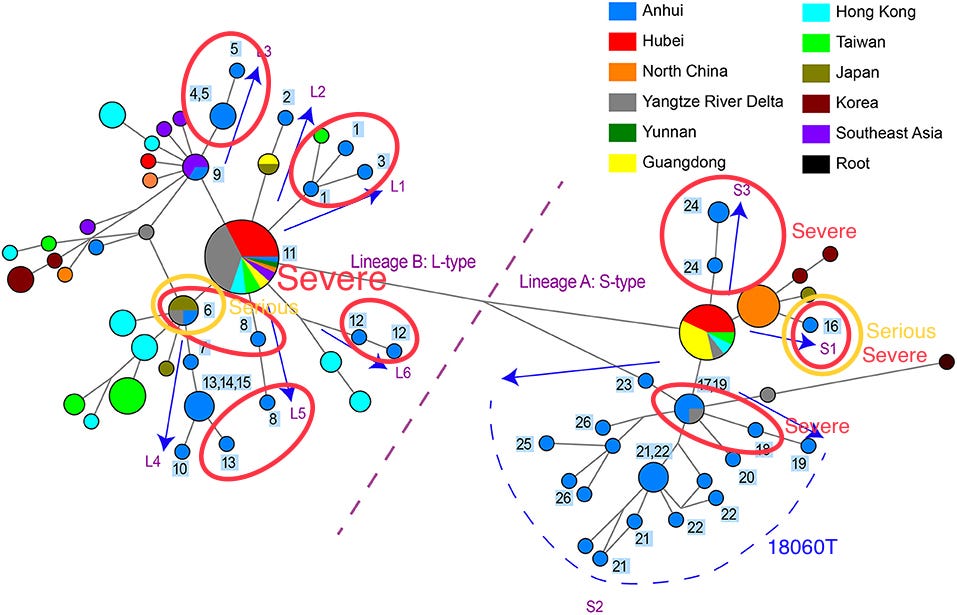
The Hefei paper, where SARS-CoV-2 genome sequences containing 18060T were reported, https://www.frontiersin.org/articles/10.3389/fmed.2021.784632/full indicated that there have been no “serious” cases (classified as having a “critical illness notice” in the medical record) in the 18060T clade, whereas the number of “severe” cases (requiring “medical intervention”) made up only 2/9 of the 18060T cases found.
Given that the ascertainment criterion in this Hefei study is “abnormality in the chest CT scan or ARDS”—this also leads to there being somewhat fewer lineage A cases sampled here compared to lineage B cases.
Analysis of patient blood test results, procured in Hefei, indicates that lineage B is statistically more likely to induce the expression of IL-6 compared to lineage A, which is more likely to induce the expression of IL-2R and SF than 18060T-carrying strains. This observed greater ability to induce more inflammatory markers within patients could be one of the causes of the higher pathogenicity of lineage B SARS-CoV-2 compared to lineage A and USA-WA-1 observed in this paper, with lineage B (8782C and 28144T) having higher severe/mild case ratio compared to lineage A (8782T and 28144C).
It has been found that IL-6 is an important mediator of inflammation,
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6801086/
Cause oxidative stress,
https://www.nature.com/articles/s41598-020-74843-x
and is known to be produced during, and enhance, an inflammatory reaction.
https://www.intechopen.com/online-first/82086
IL-6 is found to induce a viral-persistent state in several RNA virus infections, and the inflammatory state induced by IL-6 “may be an advantage for some RNA viruses by providing new targets for subsequent infections”.
The reduced expression of IL-6 associated with ORF8 84S may explain why lineage A went extinct, as this likely means that it is unable to establish a persistent infected state and can infect fewer cells within tissues compared to lineage B as it induces less inflammation, whereas inflamed tissues are better targets for RNA viruses as more target cell types within them become targets for infection. The poor ability to induce inflammation also means that lineage A is generally bad at transmitting or getting to higher titer because hyper-innate inflamed tissues are very important sites of RNA virus replication.
https://www.nature.com/articles/s41401-022-00906-6
In addition, IL-6 has been found to upregulate ACE2. This upregulation leads to more susceptible cells and higher viral titers within an infected host, which positively correlates to both viral transmissibility and symptom severity of an infection.
https://www.sciencedirect.com/science/article/pii/S0753332221012051
“Interleukin-6 IL-6 activity helps cellular entry of SARS CoV-2 virus.”
Another effect of the lower ability to induce inflammation caused by ORF8 84S is that the mutational rates for lineage A, which in coronaviruses is partially induced by stresses generated from the inflammatory response, https://pubmed.ncbi.nlm.nih.gov/15481003/
may be lower compared to that of lineage B. This creates a more recent apparent tMRCA for lineage A compared to lineage B, should a constant rate of mutation be assumed in the modeling process.
Examination of the WA1 case itself also revealed that this case failed one and half of the 4 PUE surveillance case criterions used in the Chinese NNDRS system (Fever >38C, radiographic evidence or pneumonia in lung CT image, normal/low white blood cell count and/or lymphocytopenia, and absence of improvement after 3-5 days of “standard antibiotics treatment”)—
https://www.nejm.org/doi/full/10.1056/NEJMoa2001191
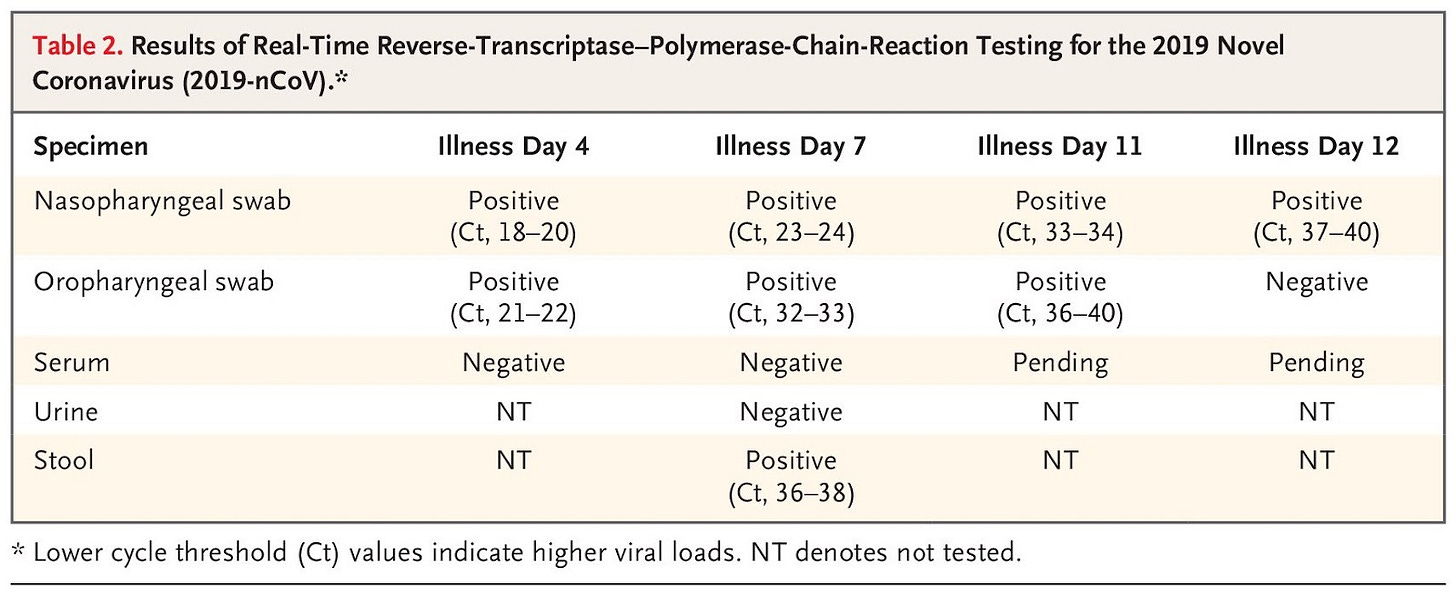
According to the case report in the article, the USA/WA-1 case began to clear the virus (swab PCR Ct value began to rise as viral titers dropped) on the 3rd day of hospitalization; and when pneumonia began to develop on the 6th day of hospitalization, was subjected to antibiotics treatment. The case recovered on the 8th day of hospitalization, which is exactly 2 days and a half after the antibiotics treatment began.
While this case also has RDV/GS-5734 use, which as a contaminant complicates the determination of antibiotics treatment result; further trials of GS-5734 found no evidence that it had any effect at reducing the viral titer of SARS-CoV-2 within patients, symptom severity, time to clinical improvement or the rate of deaths amongst treated patients.
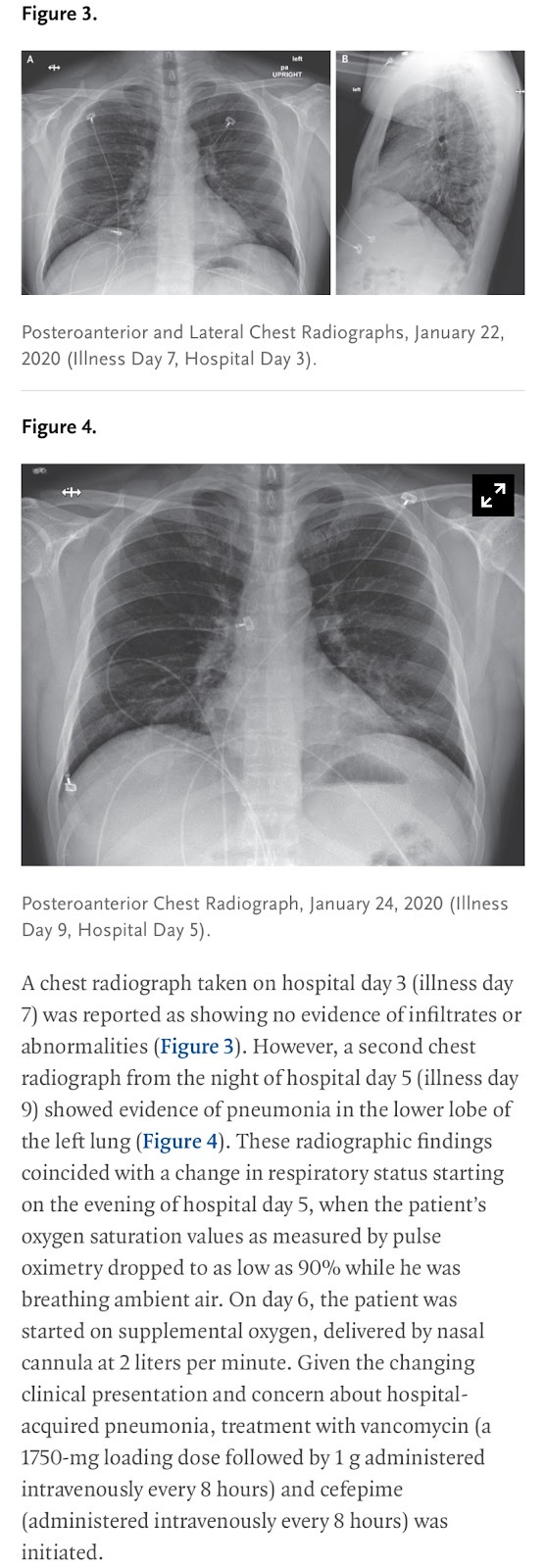
The WA1 patient is found to show leukopenia, but not lymphocytopenia. Leukocyte count rose sharply on day 7 of hospitalization as pneumonia is seen on the X-ray scan (RDV administered on “evening of day 7”, sampling time uncertain), indicating that bacterial HAP (Hospital Acquired Pneumonia) may have been contracted (neutrophil count rises with bacterial infection) on day 7 of hospitalization for the USA-WA-1 case. Residual antibiotics from the last dose on day 7 may have a role in the final resolution of the case on day 8, and it is indicated that cefepime use continued until day 8 of hospitalization for this case.
As indicated by the dropping swab PCR titer from illness day 4/7/11 and steadily increasing lymphocyte count in the same period, the patient was already recovering before the administration of RDV, and the sudden increase in neutrophil count on day 9-11 indicated that Hospital-Acquired Pneumonia had been contracted by the patient. On the “evening of day 7(hospital day 7, illness day 11)”, when RDV was administered, the swab taken (on day 7, most likely before evening) was already showing terminal viral clearing with Ct 33-34 Nasal and 36-40 Oropharyngeal—meaning that the RDV application is too late in the illness to be meaningfully affecting recovery. Antibiotics were given on day 6 of hospitalization when pneumonia signs emerged on the chest X-ray. Vancomycin and cefepime were administered—cefepime was given until day 8. Given the time needed for antibiotics and medications to work, and the course of inflammation and the resulting radiographic response (neutrophil is associated with bacteria, and inflammation/lung infiltration), it is highly likely that a 2-day course of Hospital-Acquired Pneumonia, instead of the already recovering viral infection (did not appear on X-ray before HAP), explains the condition of WA1 in the critical day 6-8 of his hospitalization.
This also raises important questions as to whether any severe enough signs of pneumonia (The Hefei paper used only “abnormal” chest radiographic/CT scan for inclusion, not specifying “pneumonia”) that warrants the beginning of CAP/HAP-associated antibiotics treatment would ever occur with WA1 without some kind of secondary infection that most likely WILL be treated and lead to recovery when the antibiotics panel was prescribed. Either viral or HAP in origin for the manifestation of the case’s symptoms, the case study of WA1 indicated that the pneumonia manifestation associated with infection of this strain of SARS-CoV-2 is too self-limiting to persist for more than 3 days when counted from the day where clinical signs would signal the beginning of antibiotics treatment for the patient.
Another interesting observation of USA-WA-1 is the apparent absence of pneumonia manifestation on Chest X-ray despite already past-peak and decreasing viral titers between illness day 3 and day 7. It is likely that unless there is a coinfecting or secondary infecting bacterium leading to HAP, signs of lung infiltrates that are sufficiently prominent to be seen on a Chest X-ray scan may not be probable with an infection with the USA-WA-1 strain.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8245302/
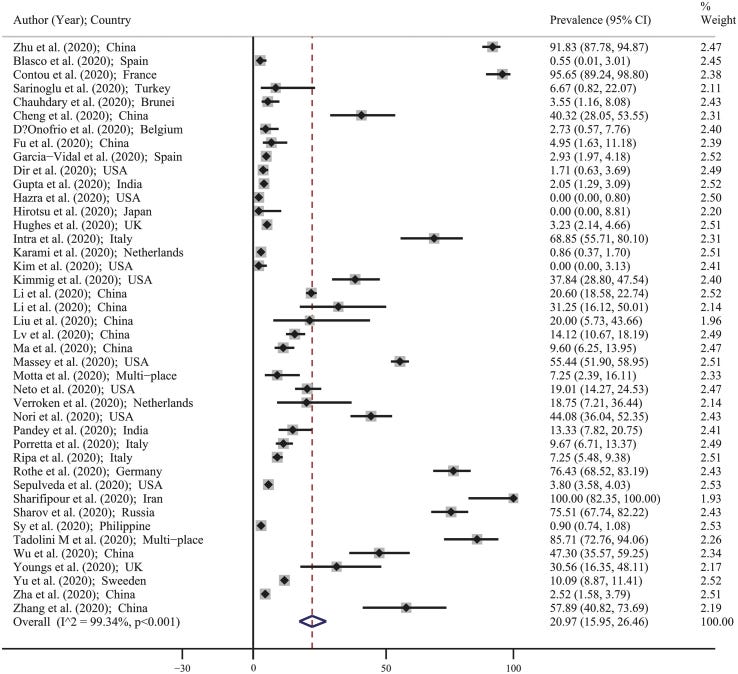
Interestingly, a meta-analysis of pooled 2020 studies indicates that the prevalence of bacterial coinfections in coarse COVID-19 patients (positive RT-PCR, before specific subtype assignment) was ~20%, where “the pooled estimate for the prevalence of bacterial coinfections was 20.97% (95% CI: 15.95–26.46). This means that overall, in every 100 people with COVID-19, 16–26 people had bacterial coinfections.”
According to the data that was used to support this result, the highest prevalence of bacterial coinfections was found in China, with “earlier” strains that are found in the initial Wuhan/China outbreak.
This is approximately the same ratio of severe patients to all patients with the allele 18060T in the Hefei paper, a ratio of 2 to 9 or 22%.
Given that it was found that https://www.accjournal.org/m/journal/view.php?number=1393
https://www.nature.com/articles/s41598-021-92220-0
bacterial coinfections correlate with higher morbidity, longer hospital stay, and substantially greater mortality even with later strains of SARS-CoV-2, this could imply that the few 18060T cases that were severe enough for medical intervention in Hefei were only so because of a bacterial coinfection/secondary HAP infection.
This means that the swift and efficient recovery following HAP onset in the USA-WA-1 sample may have been the general trend of hospitalized infections in the case of the USA-WA-1 strain.
Lastly, https://www.sciencedirect.com/science/article/pii/S0924857920305008 while ORF8 L84S (28144C) is found to be associated with milder pathogenesis in a meta-analysis, no association between mild pathogenesis and Nsp13 P504L or Y541C (17747T/17858G) were found in this meta-analysis.


As intermediate genomes between USA-WA-1 and USA-WA-2 have been found in North America, and the mutations C17747T and A17858G have not been seen in Chinese samples, it is likely that USA-WA-2 was the direct descendant of USA-WA-1 following cryptic circulation, as opposed to a separate import from China as it would require 17747T and 17858G to be found in Chinese samples, and that no intermediate genomes between USA-WA-1 and USA-WA-2 being found in North America.
In conclusion, it is likely that the presence of the USA/WA-1 lineage in Washington State is responsible for the almost nonexistent levels of excess deaths, and the absence of a significant “covid peak” in the pattern of what that did exist, most likely due to the extremely low pathogenicity of this strain compared to the A or B lineages. Analysis of the USA-WA-1 case itself indicates that this case has failed one and a half (and possibly two and a half, given that radiographic evidence of pneumonia did not show until the case contracted bacterial, hospital-acquired pneumonia on day 6 of hospitalization) of the four criteria that classify a patient as pneumonia of unknown etiology under the Chinese NNDRS system. Analysis of cases carrying 18060T in Hefei likewise gave similar results, giving the probability of such a case having “severe” outcomes being comparable to the known rate of bacterial secondary infections among COVID-19 patients in China. With neither a known link to the Huanan Seafood Market in Wuhan nor is likely to satisfy all of the four criteria for classification as a surveillance case of “pneumonia of unknown etiology”, it is unlikely that any infections caused by the USA-WA-1 strain ended up being reported or even properly hospitalized prior to the beginning of the lockdown in Wuhan. Given the duration of illness for the USA-WA-1 patient (a total of 12 days from onset to recovery), this makes it unlikely that any cases carrying 18060T would be reported in Wuhan with an onset date before 01/01/2020.






In late 2019, I had a business partner in Seattle who was entirely unproductive for most of a month. He said he had a nagging respiratory illness. For a guy in his early 30s, this didn't seem like a particularly good reason to go dark as part of a two-man team working on a startup. He never sounded like he was sick, but said he had chest congestion and a cough. The only time I heard him cough was right after he said that, and it sounded like a fake cough to assure me that he had a cough (though I believed him).
We met up in early 2020 in Denver at the Joint Mathematics Conference and I asked if he might have had COVID, and he had a weird reaction where he shook his head and declared "No" as if I'd asked if he approved of molesting kids or something.
I had to fire him in March 2020 when too much time had gone by and he was unproductive---particularly for a Stanford comp sci major with past Google credentials. I've thought back on that experience many times and think he had COVID in late 2019, but refused to accept the possibility that the narrative was wrong.
Again? Your approach is both highly detailed and conclusions as logical as a Vulcan (watch star trek to know what I mean). Excellent and very well written articulated paper. I was just 10miles from the WA1 case. I was born and raised there. Glad I moved (for many reasons).