Recently, a publication on Environmental Research provided statistically significant evidence of SARS-CoV-2 circulation in Lombardy, Italy before the first case of SARS-CoV-2 infection in the Huanan seafood market on 11/12/2019 (or officially or unofficially reported in China). This should have been sufficient to rule out the Huanan seafood market as the origin of SARS-CoV-2 for any reasonably minded person, but this is only the latest on an already growing pile of both direct and circumstantial evidence for SARS-CoV-2 circulation at least in October-November 2019.
Direct detection of SARS-CoV-2 RNA:
Two studies have reported the detection of SARS-CoV-2 RNA using PCR-based methods (including real-time qRT-PCR) and yielded results of pre-pandemic circulation of SARS-CoV-2 in a consistent time series.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7428442/
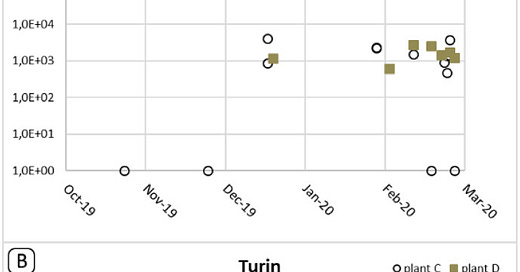
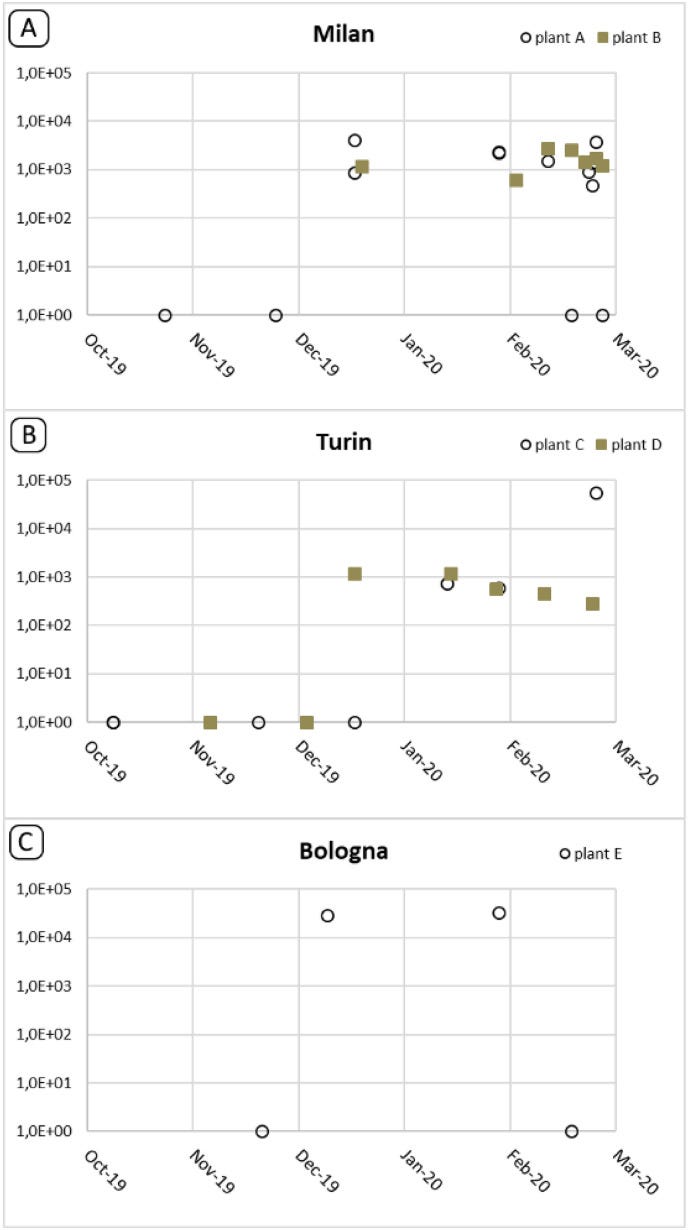
In a study of wastewater samples obtained from WWTPs (Wastewater Treatment Plants), real-time qRT-PCR assays targeting three different segments of the SARS-CoV-2 genome (E, nsp14 ExoN, RdRp) were performed using wastewater samples collected between October-December 2019 from Milan, Turin, and Bologna.
Positive results begin to show between 25/11/2019 and 18/12/2019 in Milan, between 04/12/2019 and 18/12/2019 in Turin, and between 21/11/2019 and 10/12/2019 in Bologna.
Nested PCR targeting the nsp14 ExoN CDS was performed on these samples and 12 sequences were obtained through Sanger sequencing. These sequences were found to be identical to the MN908947 reference sequence. Positivity for the nsp14 CDS was obtained at the same time as the first qRT-PCR positive samples for Milan and Turin, where positivity for this test was obtained slightly later for Bologna, on 29/01/2020.
To validate the specificity of the qRT-PCR and nested PCR tests, 24 “blank” samples obtained between September 2018 and June 2019 were tested, alongside samples of RNA obtained from other human coronaviruses, for the RNA/DNA panel of enteric viruses and bacteria (type culture obtained from EVaG). None of these samples were found to be positive for any of the 4 tests performed.
Due to the coarse temporal resolution of the sampling and testing performed in this study, The time when the level of SARS-CoV-2 circulation has reached sufficient levels to show up in the wastewater samples obtained from these 3 locations could be any time between the last negative sample and the first positive sample obtained from the WWTPs. The timeline of SARS-CoV-2 detection in these wastewater samples is consistent with what is indicated by the study of Measles-like rashes in the Lombardy region, and roughly coincides with the detection of SARS-CoV-2 RNA using PCR techniques in two other studies that were conducted in this region. As the qRT-PCR results indicate that the level of SARS-CoV-2 RNA has already peaked at the first time of detection in all 3 locations, the time when the concentration of SARS-CoV-2 reached detectable levels in these wastewater samples is likely closer to the last negative samples obtained from these regions compared to the first positive samples obtained from these regions.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7938741/
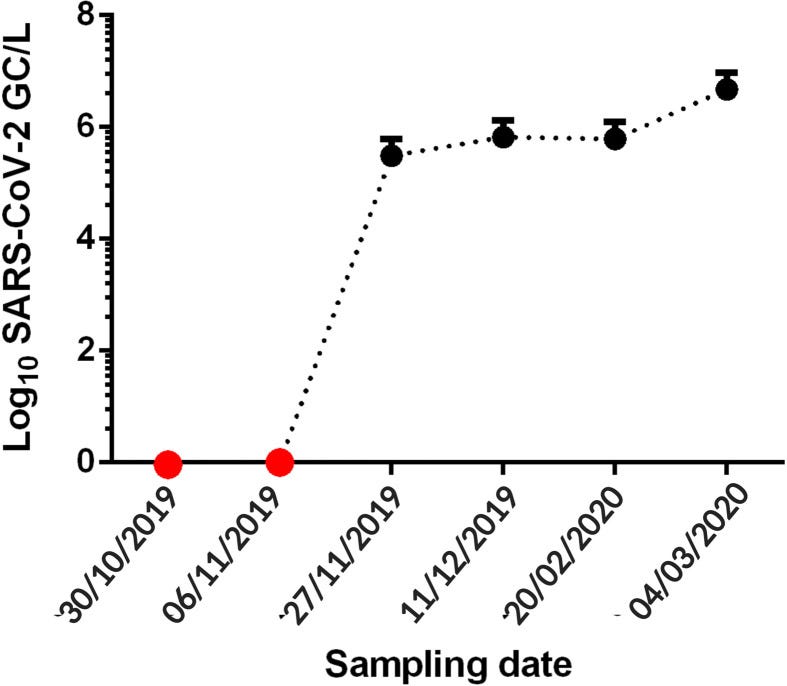
Another study producing a consistent temporal series of positive SARS-CoV-2 detection is from Santa Catalina, Brazil. This time, the SARS-CoV-2 N protein CDS, S protein CDS, and two RdRp regions were detected using the CDC-approved qRT-PCR assay on wastewater samples obtained between 30/10/2019 and 04/03/2020. a total of six raw sewage samples were collected, and testing was conducted in two independent laboratories with negative controls in the form of H2O (qRT-PCR control), as well as Field and Blank negative controls for the sampling process. All negative controls used in the test gave a negative result for the qRT-PCR test. Of the six samples that were tested in the study, the 2 samples obtained before 27/11/2019 were negative whereas all 4 samples that were obtained after 27/11/2019 were positive, with increasing titer beginning with 5.49 Log GC/L, averaging 5.83 Log GC/L and ending in 6.68 Log GC/L (Genome Copies/Liter).
Two independent NGS (next-generation sequencing) experiments were conducted on each of the two samples collected on 27/11/2019 and 04/03/2020. As mentioned in the previous post, B.1-like mutations were found while the nucleotide positions corresponding to the two miRNA binding sites in 9400-9425 and 12094-12119 were not covered in the sequencing results. By December 4, 2020, https://virological.org/t/genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-manaus-preliminary-findings/586 two highly divergent lineages, P.1 and P.2, with a B.1.28-like background and a highly divergent S protein CDS for P.1, requiring the molecular clock to be “relaxed” in order “To date the emergence of P.1, while accounting for a faster evolutionary rate along its ancestral branch”, were discovered in Brazil, with its basal lineage B.1.1.28 being a direct descendent from B.1.1 and being found mostly within Brazil. This clock anomaly strongly suggests the presence of cryptic circulation of a B.1-related lineage of SARS-CoV-2 within Brazil with a date of divergence (given a uniform molecular clock, which ratio between sampling time and divergence gives approximately 1.3~1.4 times higher divergence/perceived sample time for P.1 compared to previous B.1.1 isolates) of approximately 12 months (instead of 9 months which was made to preserve the perceived topology of the phylogenetic tree) before its discovery, landing within November 2019-January 2020.
In addition to these two major time series where SARS-CoV-2 RNA was detected in pre-pandemic samples within consistent series with a strong temporal signal, two additional simultaneous detections of SARS-CoV-2 RNA and antigens/antibodies, both in Italy, were found from 10 November 2019 and 09 December 2019.
https://www.frontiersin.org/articles/10.3389/fmicb.2022.886317/full
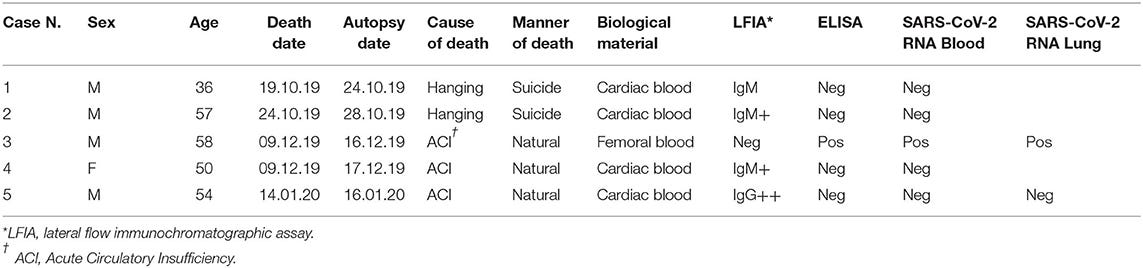
The closer one, on 09 December 2019, involves a screening of 169 autopsied patients from the city of Milan, which identified a total of five cases where a positive result of antibodies against SARS-CoV-2 was found. 4 of them contained antibodies targeting the N proteins, whereas one case, which died on 09/12/2019 due to Acute Circulatory Insufficiency, was found to contain antibodies targeting the Spike protein. When the blood and the lung tissue of the autopsied patient were tested using two different commercial quantitative PCR assays (one qRT-PCR, one droplet digital PCR(ddPCR)), both were found to be positive for SARS-CoV-2 N1 and N2 RNA, with the blood contains 38.3 RNA copies/reaction and the lung contains 289.2 RNA copies/reaction.
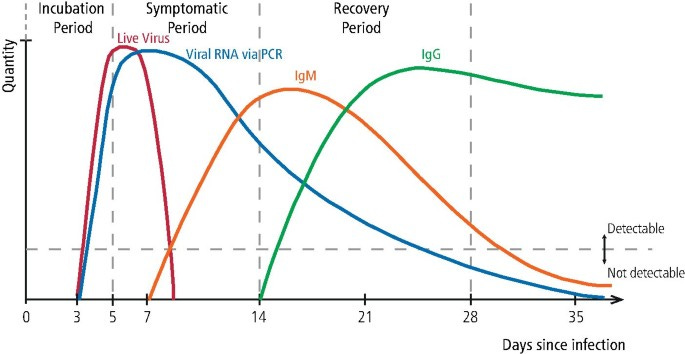
Given that the time delay from exposure to onset is 5 days, and from exposure to first antibodies (IgM) is 7 days, the latest estimated symptom onset date for this case is 07/12/2019 and was 4 days before the first official case of SARS-CoV-2 infection from China, the shrimp vendor from the Huanan seafood market with symptoms onset in 11/12/2019.
A study of patients with measles-like skin rash and morbilliform eruptions proved more fruitful.
https://onlinelibrary.wiley.com/doi/full/10.1111/bjd.19804
In another independent study, within just a single case retrospectively searched for similar symptoms from a hospital after the realization of the existence of dermatosis associated with SARS-CoV-2 infection, Immunohistochemistry staining and RNA fluorescent in-situ hybrid analysis identified both the N protein antigen and the S protein mRNA of SARS-CoV-2 (or at least a virus highly related to SARS-CoV-2 and have a Spike protein CDS sufficiently permissive for binding of the fluorescence probes to the mRNA) within the Eccrine glands of the skin biopsy sample taken in November 2019 (media reports this sample as being taken in 10/11/2019). treatment of the same sample with RNAse eliminated fluorescence, indicating the signal was generated from the interaction of the fluorescent probes with RNA within the sample. 15 other skin biopsy samples obtained in 2018 were used as negative controls while positive controls in the form of SARS-COV-2 positive patients from the pandemic era who presented with skin infections were used, with expected results from N protein-specific immunohistochemistry staining and RNA fluorescent in-situ hybridization assay. During a follow-up of the patient in June 2020, the patient was found to be positive for SARS-CoV-2 specific IgG after the resolution of dermatosis in April 2020.
While the authors were unable to detect amplifiable SARS-CoV-2 nucleic acids using RT-PCR-based methods, this is not unusual for formalin-fixed and paraffin-embedded samples as without specialized extraction and sample preparation protocols to first remove modifications and restore the template activity of the RNA, RNA extracted from formalin-fixed and paraffin-embedded samples are unusable as RT-PCR template and amplification was found to be difficult and inconsistent, with most reactions not able to produce amplification at all while others were only able to generate very weak levels of amplification. In contrast, formalin-fixed and paraffin-embedded samples are suitable RNA-FISH substrates for the detection of a variety of pathogens in tissue samples.
Detection of SARS-CoV-2 antigens and antibodies:
One significant detection of SARS-CoV-2 antigens and one consistent temporal series of SARS-CoV-2 antibodies were found in Boston, Massachusetts, and in France.
Boston: “upper respiratory infections, bacterial pneumonia, viral pneumonia, or unspecified virus positive”?
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7499543/
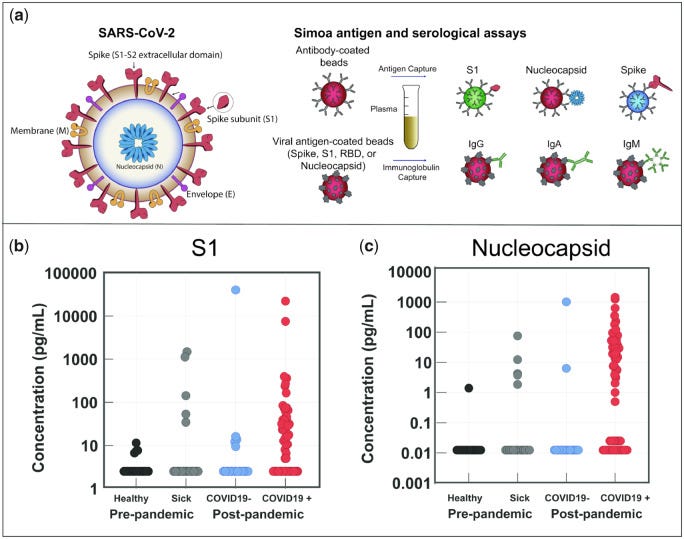
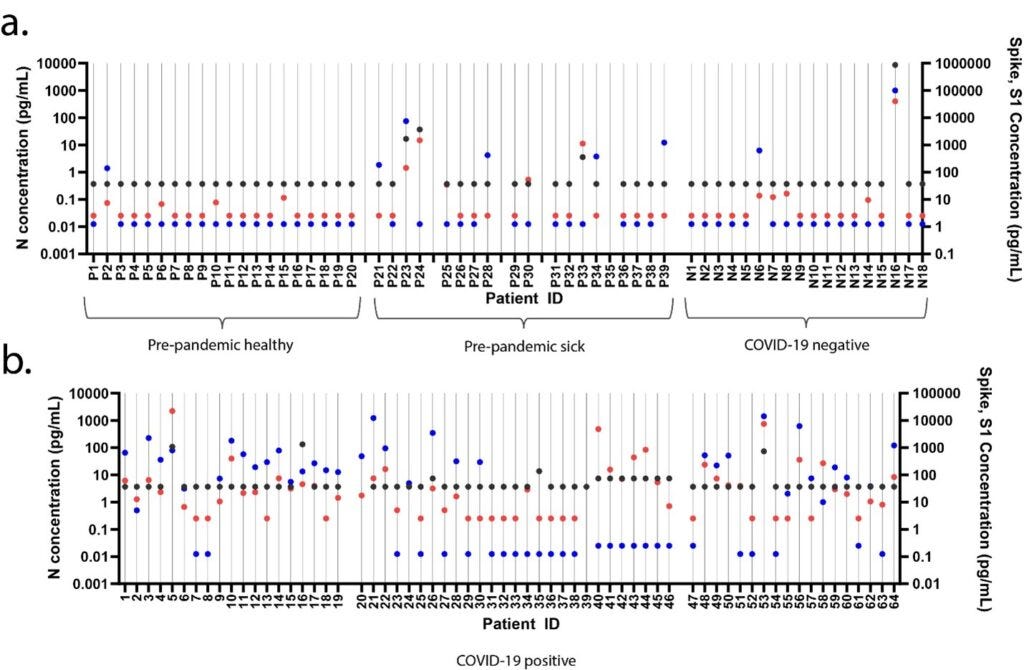
The first detection of SARS-COV-2-specific antigens came with a bit of surprise: rather than trying to find evidence of pre-pandemic circulation for SARS-CoV-2, the authors of the study were instead developing a highly-sensitivity antigen-based assay in order to detect the presence of SARS-CoV-2 antigens within patient samples. monoclonal antibodies were first generated against the S1 protein of SARS-CoV-2 through the use of phage display, then a Simoa immune assay using this S1-specific monoclonal antibody as well as commercial anti-S and anti-N antibodies to detect the presence of viral antigens within different cohorts of patients: patient samples collected before October 2019, that were healthy collection. patient samples collected before October 2019, that have “upper respiratory infections, bacterial pneumonia, viral pneumonia, or unspecified virus positive” at collection. Patient samples collected during the pandemic, and tested negative for SARS-CoV-2 RNA on a qRT-PCR test. Patient samples collected during the pandemic, and tested positive for SARS-CoV-2 RNA on a qRT-PCR test.
Despite the high specificity of the S1 antigen during an immunological assay for SARS-CoV-2, instead of finding all negative results in the pre-pandemic cohort, the authors were surprised that 3 out of the 14 sick pre-pandemic patients contained a concentration of the S1 protein above 100pg/ml, whereas the highest level of S1 protein antigens within the pre-pandemic healthy cohort was only about 10pg/ml. None of the pre-pandemic healthy cohort patients contained a simultaneous positive detection of S and S1, whereas one of the patients in the pre-pandemic sick cohort and another patient in the pandemic SARS-CoV-2 PCR negative cohort was found to simultaneously have both the S, S1, and N antigens (likely due to false negative qRT-PCR for the pandemic patient). It was found that the level of S1 and N detections in the pre-pandemic sick cohort was higher than both the pre-pandemic healthy cohort and the pandemic SARS-CoV-2 PCR negative cohort, indicating that a significant signal for the presence of SARS-CoV-2 related antigens within sick patients with respiratory illness in Boston at least as early as October 2019.
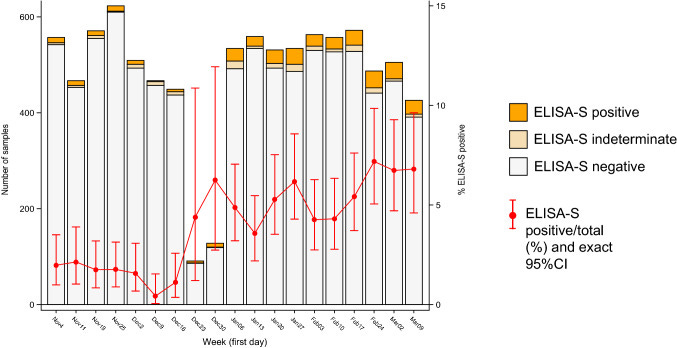
France: evidence of population-level IgG seroconversion during mid-December 2019.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7864798/
Series of samples indicating the pre-pandemic circulation of SARS-CoV-2 exist not only in RNA samples but also in serological (antibody) samples as well. During a nationwide retrospective analysis of blood serum samples from 12 main regions in France, SARS-CoV-2 Spike-reactive IgG antibodies were found within patient samples collected between 04 November 2019 and 09 March 2020.
A sudden increase in the percentage of samples positive for SARS-CoV-2 Spike-specific IgG antibodies was found over the course of 16 December 2019 to 30 December 2019. France observed the first sample positive for SARS-CoV-2 RNA in a targeted search in the ICU (Intense Care Units) in a hospital in Paris on 27 December 2019. This case did not have any reported foreign travel history after August 2019, to Algeria.
As IgG antibodies usually take about 14 days or two weeks post-exposure to develop for SARS-CoV-2, a seroconversion series for IgG antibodies beginning in the 16/12/2019-22/12/2019 period would imply the beginning of significant community-level SARS-CoV-2 exposure between 02/12/2019 and 09/12/2019. This once again indicates that the global circulation of SARS-CoV-2 have already begun before the first officially announced case of SARS-CoV-2
Apart from these molecular detections of SARS-CoV-2 RNA, antigens, or antibodies before the first officially reported cases of SARS-CoV-2 at the Huanan seafood market, circumstantial evidence of SARS-CoV-2 related illnesses and anomalies are found all over the world in the second half of the year 2019.
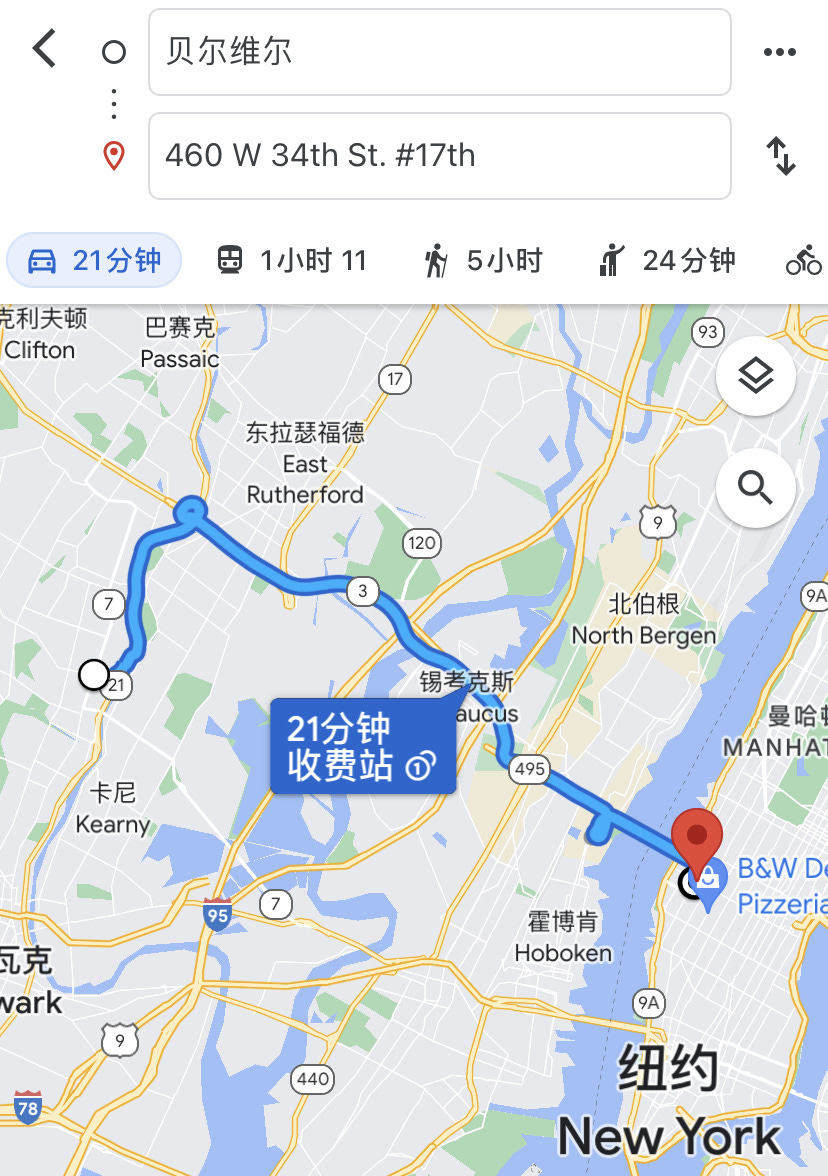
New York: Mayor recalls symptoms in November, test positive for antibodies
On April 30, 2020, the mayor of Belleville, Michael Melham, announced that he has tested positive for antibodies targeting SARS-CoV-2 while recalling symptoms that are highly consistent with SARS-CoV-2 infection on November 21. 2019.
He reports first “feeling awful, but writing it off as a mix of exhaustion and dehydration from the three-day event.”, then “Friday and Saturday of that week were no better. By Sunday night, Melham was awake all night, battling chills, hallucinations and a skyrocketing temperature.”. Initially suspected as “a case of the flu and told him that he’d recover with a few days of rest”, Michael Melham would only become better “eventually”, after at least four days or possibly later. Suspicion eventually drove him to take a blood test for SARS-CoV-2 antibodies, which then came back positive.
Florida: First reported COVID-19 death in 11/01/2020
Despite the official claim that the first case of SARS-CoV-2 infection in the U.S. is from a patient returning from Wuhan on 15 January 2020, in the three months following this first report, The National Center for Health Statistics has seen within its dataset for “all deaths involving COVID-19” with “week ending date in which the death occurred” being filled in with death reports happening as early as 11/01/2020. After a set of previously disappeared statistical data about COVID-19 cases in Florida have returned to public view, about 171 officially registered COVID-19 cases within Florida have been found to “had experienced symptoms in January and February, long before state officials acknowledged the spread of the disease”. A further examination of the dataset has revealed that “More than 60 percent of them reported no out-of-state travel. None reported travel to China.”, whereas the reason for them being reported so late was that “In January, the U.S. Centers for Disease Control and Prevention would only test for the virus if the patient had been in China or been in close contact with such a traveler.”.
While it is likely that death certifiers were retrospectively filling the cause of death for several cases that died as early as 11/01/2020 as “COVID-19” in the United States, the early requirement for contact or travel history to China may have left much of the population untested and missed many earlier cases.
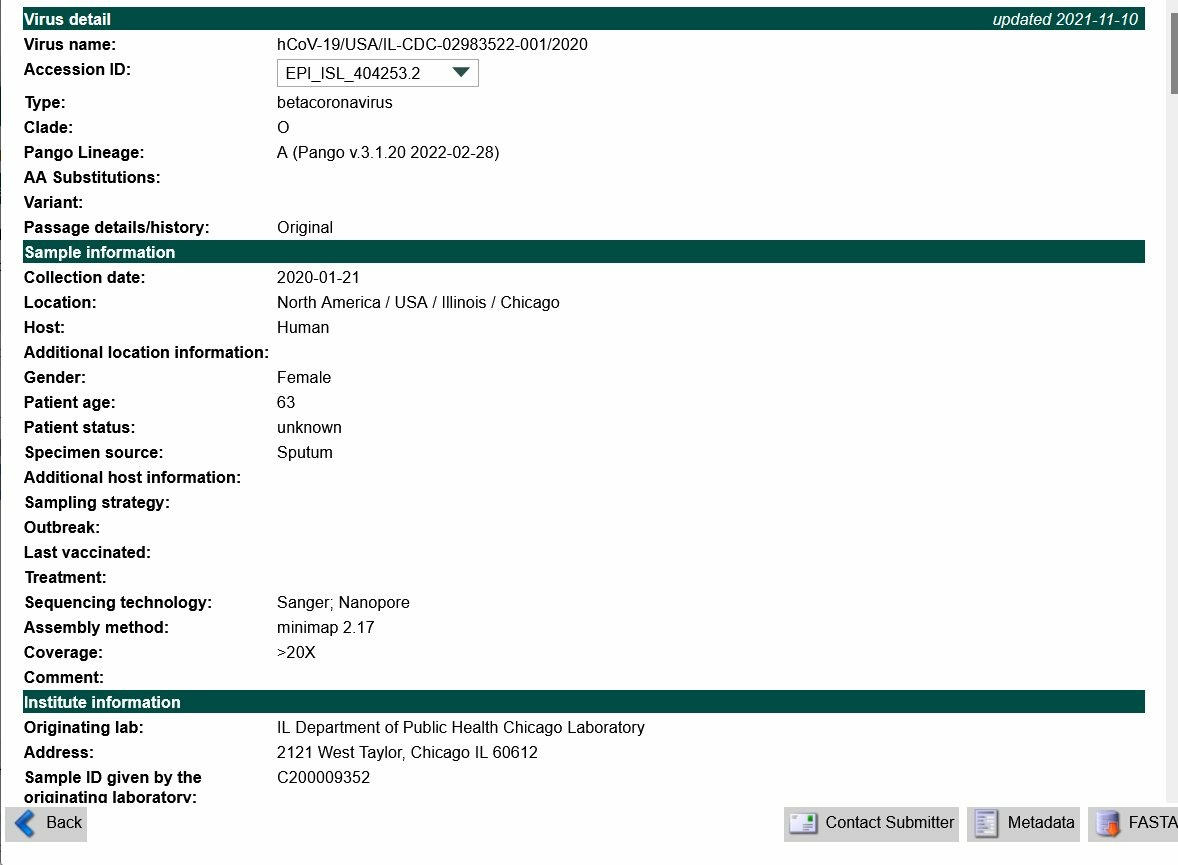
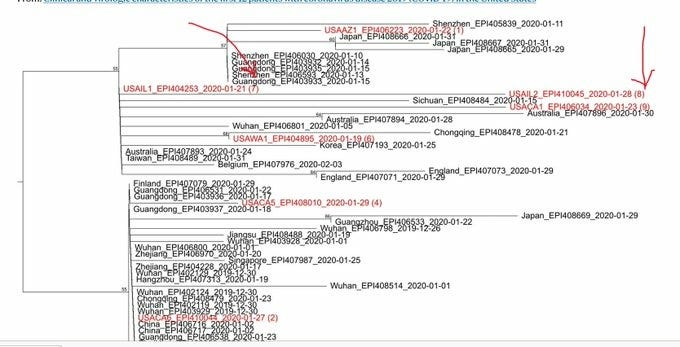
Illinois: “Family cluster” of “travel-related COVID-19 cases” with distant genomes suggestive of community transmission instead of contact transmission.
In the publication supposedly documenting the first human-to-human transmission of SARS-CoV-2 in the U.S., The genomes for SARS-CoV-2 isolated from the two cases were EPI_ISL_404253 and EPI_ISL_410045. However, closer examination of the 2 genomes revealed that the two genomes were at least 3AA different from each other, which places a low probability that the two genomes were generated from a single human-to-human transmission event from one another. This indicates that significant community transmission of SARS-CoV-2 has already begun in Illinois at the first official report of human-to-human transmission of SARS-COV-2 on 23 Jan 2020 and 30 Jan 2020.
https://www.thelancet.com/journals/lancet/article/PIIS0140-6736(20)30607-3/fulltext
Iran: Three Caspian Sea provinces begin to show excess deaths in fall 2019
https://www.sciencedirect.com/science/article/pii/S120197122100326X#fig0005
Despite attempting to defend the claim that “COVID-19 did not start in Iran until December 2019” using nationwide excess mortality data, the curve of excess mortality within three provinces in Iran, Gilan, Golestan and Mazandaran, were found to be consistently above past ranges since the fall of 2019 (September-December 2019). With the exception of Gilan where excess mortality begins to decrease in Spring-Summer 2020 (March-September 2020), Golestan and Mazandaran continues their excess mortality trend into the pandemic as the three provinces (especially Mazandaran) would become the hardest hit provinces by COVID-19 in Iran at the beginning of the pandemic.
Alongside the province of Tehran, Qom, and Markazi, where an increasing trend of excess deaths begin to show in Fall 2019, Winter 2019, and Spring 2020, The provinces that begins to show a consistently increasing trend of excess deaths starting in fall 2019 were all located nearby the Caspian Sea. This area is known to be one of the locations where U.S. troop deployment and supply chains go through or are located near, particularly since the year 2018. Golestan is found to border the capital city of Turkmenistan, Ashgabat, and the USAID and “Peace Corps”, a program run by the United States and participated mostly by American Citizens. Programs of the USAID trialing “new TB drug treatment” were active in 2019. Gilan borders Azerbaijan, a location with a known Biolab that was funded by the U.S. A program, named “Bats for peace”, was found to be running in Georgia, in the period between 2018-2019.
More recently, two SARS-CoV-2 genomes carrying an RBD position ancestrally conserved in ACE2-using Sarbecoviruses, Q498H(Y/F for SARS-COV related viruses), were found in Iran. These 2 genomes are EPI_ISL_672581 and EPI_ISL_672589.
Ottawa: dead bodies “pile up so much that the morgue was overflowing” in 2019
In an early news release on January 07, 2020, The Ottawa Hospital in Canada reported:
““During an extraordinary period in December, it was necessary to place deceased individuals outside of the designated morgue, in a secure room, for a short period of time,” it read.
“The Ottawa Hospital has converted spaces, formerly used for autopsies, within the morgue to manage unexpected surges in demand. Conference rooms and ward beds are not used for housing deceased individuals,” the statement continued.””
An interesting result from a study of SARS-CoV-2 positive cases from the first wave of the recognized SARS-CoV-2 outbreak in the Canadian state of Quebec indicated that “There was very little reported travel from Asia (n = 4, 1.2%) and none from China.” This is at odds with the fact that ~89400 Chinese nationals lived in Quebec. However, a similar discovery was found in the first pandemic wave in Italy, where it was found that in the town of Prato, a location with a 10% Chinese population, “Not a single member of the Chinese community there has tested positive for the virus” in the first pandemic wave in Italy.
Fairfax county: Respiratory illness where “Despite extensive testing of multiple specimens, no specific pathogen was identified as the cause of the outbreak.”
On July 11, 2019, an unusual outbreak of “respiratory illness” happened in a retirement care facility in Fairfax, Virginia. The outbreak affected a total of 63 patients with symptoms from upper respiratory symptoms to pneumonia. 23 patients were hospitalized and two have died.
Testing of the outbreak cases was conducted by the local CDC, but the results given the posted updates have been inconsistent across updates. For the first 17 samples sent to the CDC on July 17, 2019, no specific cause of the outbreak was identified. CDC testing of samples from July 19, 2019, indicated that some of the samples (“several samples”) were positive for rhinovirus, while other samples contained only communal bacteria that “may not be the cause of infection”. Despite several samples testing positive for rhinovirus, on July 26, 2019, the CDC testing result report indicated that “Despite extensive testing of multiple specimens, no specific pathogen was identified as the cause of the outbreak.” On 29 December 2019, despite evidently samples have already been tested with positive rhinovirus result not being seen in all of the samples sent for testing, “Results of earlier testing submitted to the Centers for Disease Control and Prevention indicated rhinovirus, a virus that causes the common cold.” were given. However, the exact number of samples where rhinovirus has been found or whether this is the case for all of the samples that were collected and sent during the outbreak were not given. There is no mention of H.influenzae in these July 2019 reports or the bacterial testing results indicated in the reports.
As of 11/05/2020, the Fairfax County government has refused the call to re-investigate samples from the July outbreak for SARS-CoV-2 infection, claiming “There is no need to re-investigate the outbreak”. The exact prevalence of either H.influenzae or Rhinovirus was not given, nor if these 2 pathogens were found in all samples collected from the Fairfax outbreak.
Rhinovirus was observed in 3.6% of general patients that tested positive for SARS-CoV-2 in a meta-analysis and H.influenzae was observed in 6.6% of such patients. However, in some locations, the prevalence can raise to as high as 31.9% for Rhinovirus and 17.2% for H.influenzae.
About a month after the Fairfax outbreak, The USAMRIID laboratory complex in Ft.detrick was shut down due to a lack of “sufficient systems in place to decontaminate wastewater”.
Maryland: “Only four of the 32 cases sent to state labs tested positive”
Just after the outbreak of respiratory illness in Fairfax, Virginia on July 17, 2019, another outbreak of “more than 3,000 sneezing, coughing, miserable people with flu-like symptoms so far” on 30 October 2019. In addition to being ~3 weeks earlier than the usual beginning of the flu season ILI spike across 2014-2019, “But not all of the 3,154 cases seen in Maryland were actually influenza. According to the Maryland Department of Health’s weekly flu report, 111 cases responded positive to rapid flu tests. Only four of the 32 cases sent to state labs tested positive.”
Interestingly, “A spokeswoman for Frederick Health Hospital did not return a request for comment on any cases seen in the emergency department.” and “the Maryland Department of Health does not release the location of each case”.
National ILI surveillance in the U.S.: Excess ILI cases begun cropping up in November 2019
http://web.archive.org/web/20210126194739/https://www.cdc.gov/flu/weekly/weeklyarchives2020-2021/ILI02.html
In the last 8 weeks of 2019, the United States National influenza-like illness surveillance network has begun to report an above-baseline level of Influenza-like illness approximately 3 weeks before the beginning of an uptick of such illnesses since the 2009-2010 H1N1 pandemic. High levels of influenza-like illness reports will continue until the first wave of recognized COVID-19 begins to raise in March-April 2020 when ILI case reports began to be recognized and registered instead as COVID-19 case reports.
Britain: Patient with onset on “15 December 2019”, no travel history, died with SARS-CoV-2 in the lungs
Initially sick with a fever and cough on 15 December 2019, an 89-year-old retired company secretary from Chatham, Kent, was hospitalized on January 7 2020 as his cough gradually worsened. His daughter began showing “symptoms of the virus” “including a dry cough, and fever, as well as aches and pains and diarrhea” “before Christmas”, whereas his granddaughter began to show a cough and fever on January 10, 2020. The official first case of SARS-CoV-2 infection in the U.K. were two Chinese nationals, who arrived on 23/01/2020 and fell ill on 26/01/2020 in York, 16 days after the Kent patient have been hospitalized. The patient eventually died on 30 January 2020, whereas when the lung tissue of the patient was tested, “tests carried out after his death revealed Covid-19 was present in his lung tissue”. The distance from York to Kent was 246.9 miles, and the 2 official first cases were quickly quarantined on 28/01/2020 as the younger of the 2 official UK index cases (the son of the older case) fell ill on 28/01/2020. Given the apparent serial interval from arrival to the onset of the son on 28/01/2020 of ~2 (by symptoms onset) to 5 (by contact) days, with a more comprehensive meta-analysis giving 4.2 to 7.5 days, and given the hospitalized (and therefore, lack of contact) nature of the 89-year-old granddad in Kent, community transmission emanating from these 2 official index cases is unlikely to reach into Kent and create the positivity within this patient, within at most 1 week of available time, without first causing other symptomatic infections in its way through London. None of the family members of the 89-year-old Kent cases (where two fell ill with COVID-19 compatible symptoms before the date of arrival of the first imported COVID-19 case in the U.K.) had travel history outside the U.K., making the date of the beginning for the community spread that infected this family (especially the granddad) comparable to or slightly earlier than the first official Chinese COVID-19 case with symptoms onset on 11 December 2019. While one of the daughters attended a “Christmas party”, this party was in December 2019 and foreign nationals were not indicated to be in the party. This U.K. family cluster thus represents another highly likely evidence of community transmission beginning outside China before the first case of SARS-CoV-2 infection at the Huanan seafood market.












I'm late to your post here, came from Dr. Mengelefaucin koronalinko replying to a comment I made on - oops not sure where I was. He gave a list of articles and yours was there. Fascinating. Also, the Ft. Detrick/Fairfax County coinciditis stood out to me as rather damning.
Thank you for putting this together! I'm passing it on to others.
So glad you decided to start up your own Substack!